Figure 4.
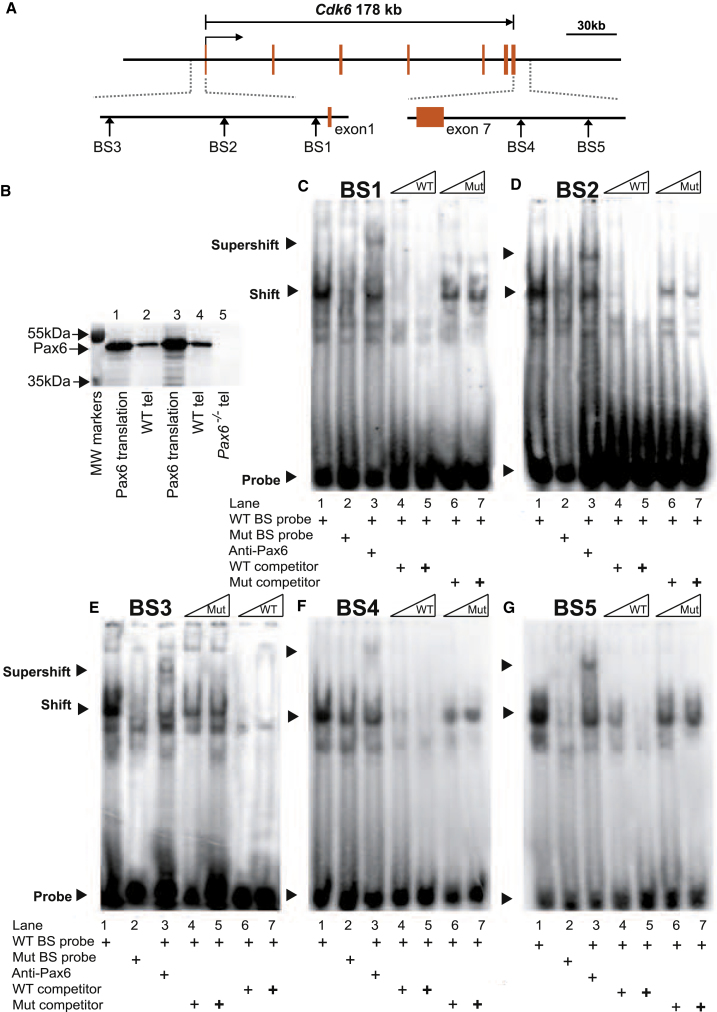
EMSAs Show that Pax6 Can Bind to Predicted Binding Sites (BS1–BS5) at the Cdk6 Locus
(A) Map of the five binding sites (BS1–BS5) at the Cdk6 locus (sequences in Figure S6A).
(B) Western blot analysis of Pax6 protein expressed by the TNT In Vitro Transcription/Translation System. Lanes were loaded with 2 μl (lane 1) and 4 μl (lane 3) of in vitro-translated Pax6 protein, 20 μg (lane 2) and 40 μg (lane 4) of Pax6+/+ telencephalic protein, or 40 μg of Pax6−/− telencephalic protein (lane 5).
(C–G) EMSAs. Arrowheads indicate free probe (Probe) at the bottom of each gel (probe is WT in lanes 1 and 3–7, and mutant (Mut) in lane 2), probe-protein complexes (Shift), and probe-protein-antibody complexes (Supershift). In all cases, a specific gel shift by binding of Pax6 to potential Pax6 binding sites was clearly detected (lane 1) and was greatly reduced or abolished by mutation of the Pax6 binding sites (lane 2). Binding specificity was confirmed by preincubation with anti-Pax6 antibody, resulting in supershifted complexes (lane 3). Visible shifts were diminished or abolished (in a dose-dependent manner in most cases) by addition of excess nonradiolabeled WT probes to compete with radiolabeled probes, whereas excess amounts of nonradiolabeled mutant probes competed much less effectively (lanes 4–7).
See also Figure S6A.