Figure 2.
ccdc111 Is a DNA-Dependent DNA Polymerase
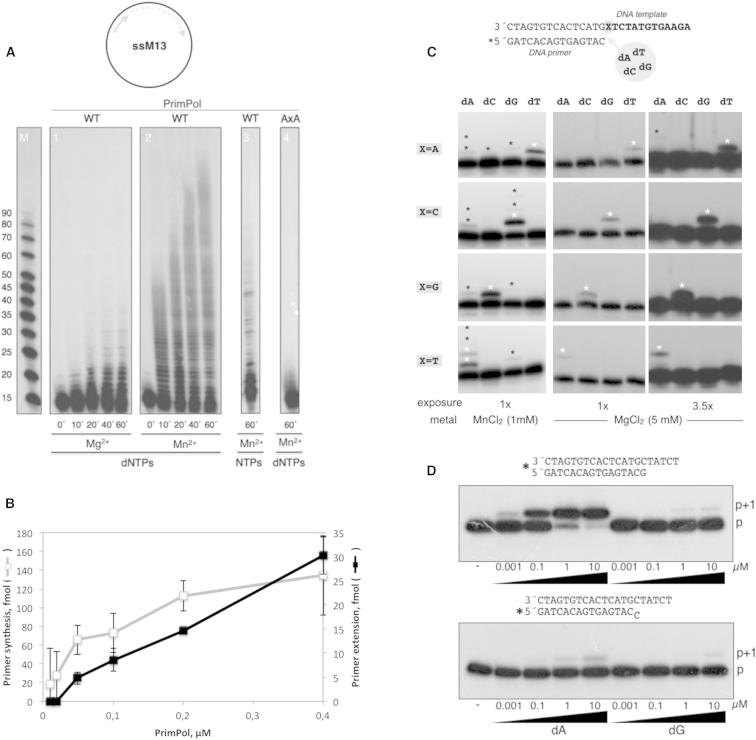
(A) Elongation of a 17-mer primer annealed to M13 circular ssDNA in the presence of either 5 mM MgCl2 or 1 mM MnCl2 as metal activators. WT ccdc111 (but not mutant AxA) extended the primer producing elongation intermediates characteristic of distributive DNA synthesis. NTPs were worse substrates for polymerization by ccdc111.
(B) Compared efficiency of primase and polymerase activities of PrimPol. The primase assay, using the 29-mer GTCC oligo (5′-T15CCTGT10-3′) as template, and the DNA polymerase assay, using a template/primer oligonucleotide, were carried out as described in the Experimental Procedures. Data are represented as mean ± SD (n = 3). See also Figure S3.
(C) Matched versus mismatched nucleotide insertion at the four template bases. The four template/primer structures used, differing in the first template base (X), are indicated. Nucleotide insertion on each template/primer was analyzed in the presence of each individual dNTP. PrimPol preferentially inserted the complementary nucleotide dictated by the first available templating base. The insertion of some errors (indicated with asterisks) can be detected.
(D) Polymerization assays were carried out with either a matched (top) or mismatched (middle) template/primer and the indicated concentrations of a correct (dA) or wrong (dG) nucleotide. After incubation, +1 extension of the 5′-labeled (∗) strand was analyzed by denaturing gel and autoradiography.