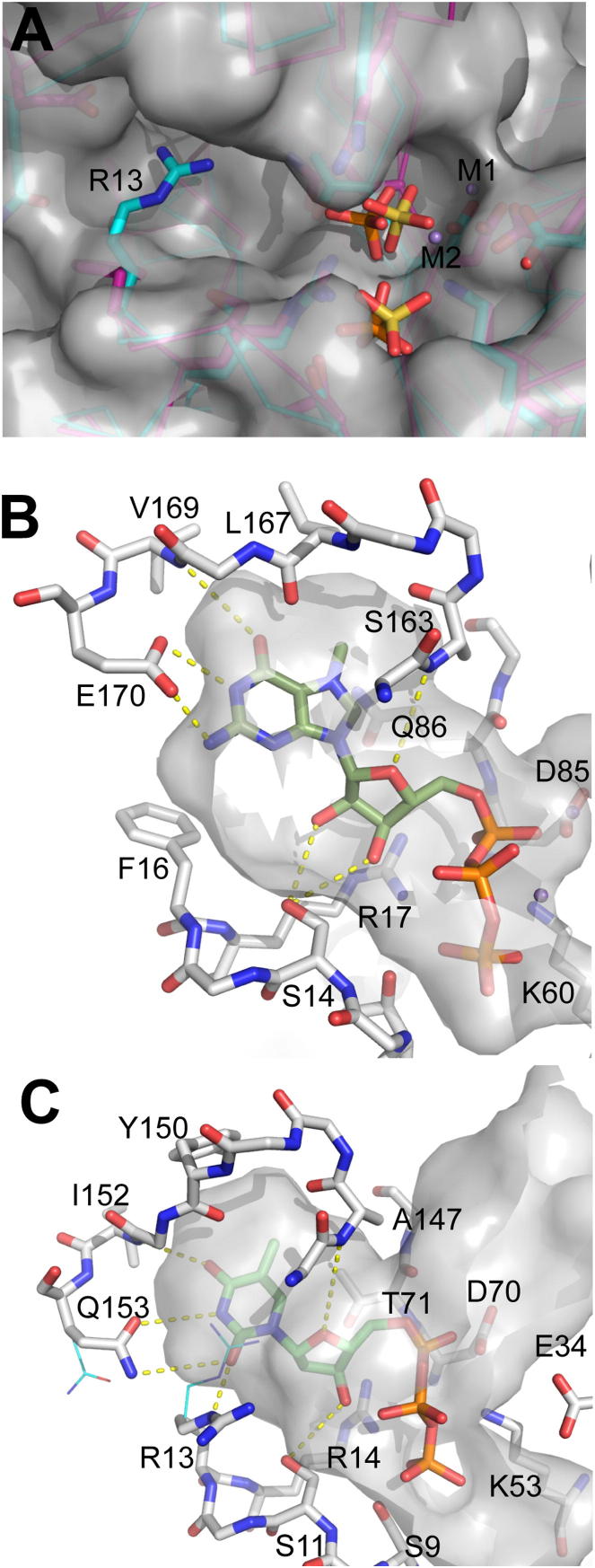
Figure 6.
Structural Basis of Different Substrate Specificity of Two Maf Subfamilies
(A) Structural superposition of the substrate binding sites of BSU28050 (cyan; YhdE subfamily) and Tb-Maf1 (magenta; YceF subfamily). Side chains of conserved residues, as well as phosphate (orange) and sulfate (yellow) ions bound to BSU28050 and Tb-Maf1, respectively, are shown as sticks. Also shown are the bound Mn2+ ions (M1 and M2) and the surface of the Tb-Maf1. In BSU28050, the side chain of the conserved Arg13 is located in the base-binding pocket, but it can be flipped away without altering the main chain conformation.
(B) The base-binding pocket of the Tb-Maf1 (YceF subfamily) with manually docked m7GTP (dark green carbons). The predicted substrate specificity determinants are the Val173 main chain amide, Gln90, and Glu174 (labeled and also shown with potential H-bonds).
(C) The base-binding pocket of the BSU28050 (YhdE subfamily) with manually docked dTTP (light green carbons). The predicted substrate specificity determinants are Arg13, Thr71, and Gln153 (labeled and also shown with potential H-bonds). The side chain conformations of Arg13 and Gln153 have been changed to different rotamers compared to the original structure shown in (A).
See also Figure S7.