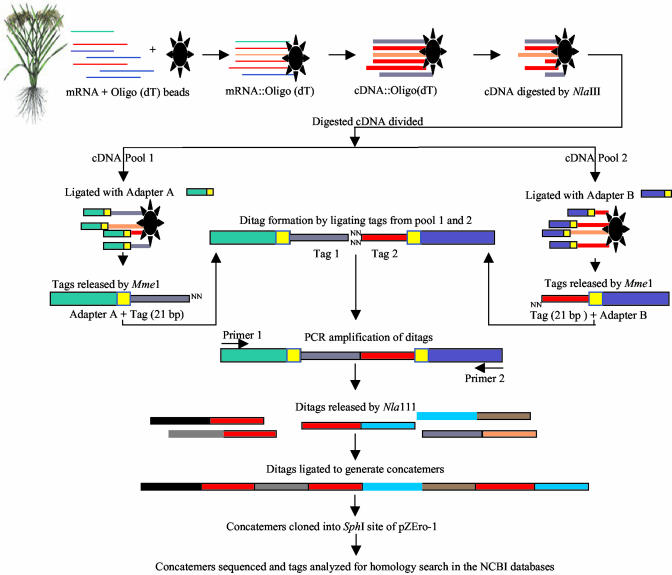
Figure 1.
Schematic representation of LongSAGE technology based on Saha et al. (2002). Total RNA was isolated from rice leaves, and mRNAs was purified and covalently linked to oligo(dT)25 magnetic beads. Single- and double-stranded cDNA was synthesized and digested with NlaIII. The digested cDNA was equally divided into two parts, pool 1 and 2, and ligated with linkers A and B, respectively. These linkers consist of an asymmetric recognition motif for a type IIS enzyme, MmeI, and specific sequences for PCR primer-binding sites. The ligated cassettes (linker::cDNA) were treated with MmeI to release tags from cDNA. The MmeI tags (LongSAGE tags) from pools 1 and 2 were mixed together (sticky end ligation) to form ditag cassettes. These cassettes were PCR amplified using primers specific to each linkers. Then linker sequences were removed from the cassettes by NlaIII digestion. Ditags were ligated together to generate longer molecules called concatemers. Over 0.5-kb concatemers were cloned into SphI site of pZEro-1 and sequenced using M13 primers. Ditags and tags were extracted from a high quality sequence data and homology search was conducted using the National Center for Biotechnology Information (NCBI) databases.