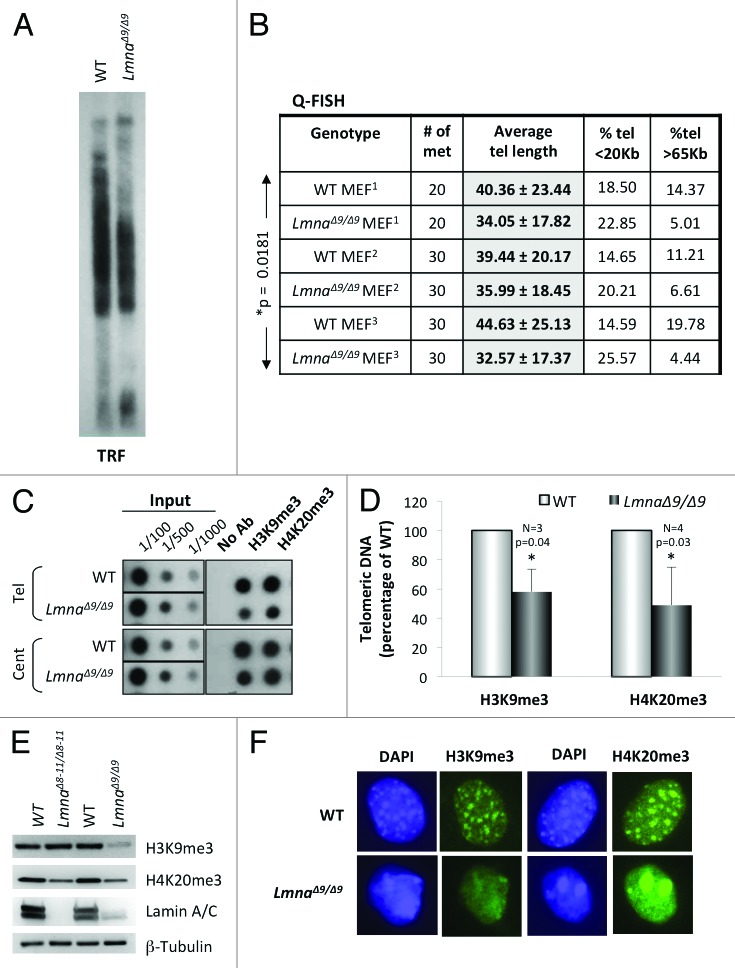
Figure 1.Telomere shortening and heterochromatin defects in LmnaΔ9/Δ9 MEFs. (A) Telomere restriction fragment (TRF) analysis shows faster migration of telomeres and thus telomere shortening in LmnaΔ9/Δ9 MEFs. (B) Three independent Q-FISH analyses of telomere length in wild-type and LmnaΔ9/Δ9 MEFs. Telomere length of at least 20 metaphases was measured per experiment. Note the decreased average telomere length in LmnaΔ9/Δ9 fibroblasts. (C) ChIP analysis performed on wild-type and LmnaΔ9/Δ9 MEFs with antibodies recognizing H3K9me3 and H4K20me3. Immunoprecipitated DNA was dot blotted and hybridized to a telomere probe, stripped and rehybridized to a centromere probe (major satellite). (D) Quantitation of immunoprecipitated telomeric DNA after normalization to input signals with H3K9me3 (3 experiments) or H4K20me3 (4 experiments). Bars represent standard deviation. (E) Western blots to monitor global levels of H3K9me3 and H4K20me3 in wild-type, LmnaΔ8–11/Δ8–11 and LmnaΔ9/Δ9 MEFs. β-Tubulin was used as loading control. (F) Immunofluorescence performed in wild-type and LmnaΔ9/Δ9 MEFs with antibodies recognizing H3K9me3 and H4K20me3 (green). DAPI is shown in blue. Note the diffuse and irregular distribution of heterochromatin domains in the mutant cells. *P, represents value of statistical significance; P ≤ 0.05 is considered significant.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.