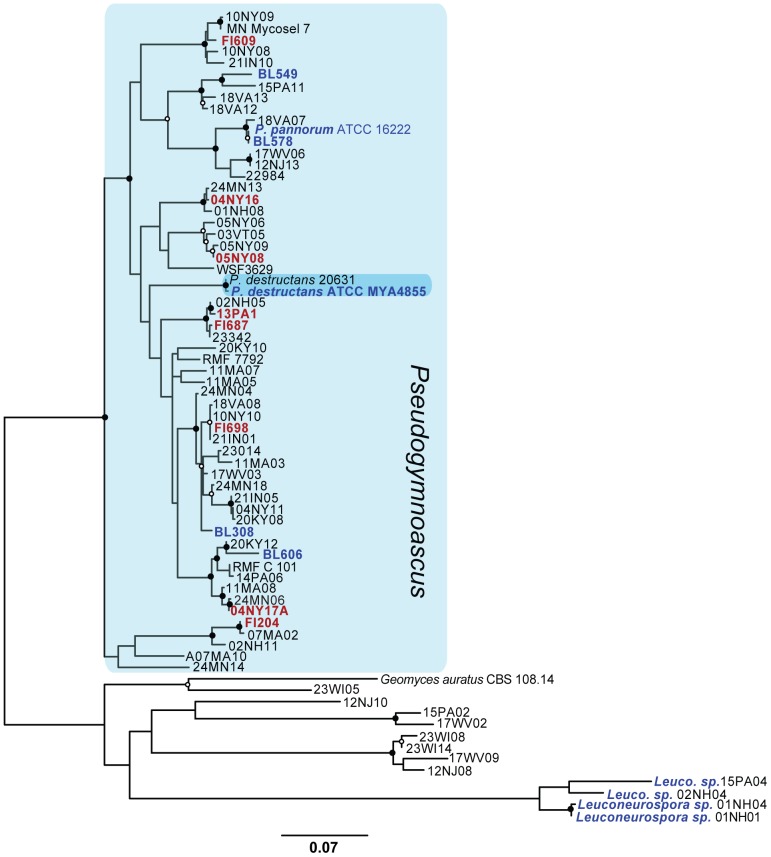
Figure 1. Best maximum-likelihood tree for the Pseudogymnoascus spp. used in this study (lnL = −10622.006982).
A concatenated alignment of the TEF-1, MCM7, and partial ITS genes was used to generate a Maximum-likelihood tree in RAxML using strains identified in Minnis and Lindner [5]. Values show bootstrap support from 500 bootstrap replicates; closed circles indicate ≥95% support at each branch point, while open circles indicate >70% support. The taxa shown in red were used in 10X tests, while the taxa in blue were used in triplicate tests. Geomyces auratus and members of the Leuconeurospora were used as the outgroups. Accession numbers for each sequence can be found in Tables 1 and S1.