Abstract
A search of the recently completed genomic database of rice (Oryza sativa) identified a 29-member xyloglucan endotransglucosylase/hydrolase (OsXTH) gene family. This first report of a complete XTH family from a monocotyledonous species reveals that the OsXTH family is comparable in size with that of the dicotyledon Arabidopsis thaliana, which consists of 33 AtXTH genes. This is surprising because xyloglucan, the specific substrate of XTHs, is considerably less abundant in cell walls of monocotyledons than dicotyledons and is not typically ascribed an important structural role in monocotyledons. As a first step toward determining the roles of rice XTHs, the expression patterns of all 29 OsXTH genes were examined using a quantitative DNA microarray procedure with gene-specific oligonucleotide probes. The analysis showed that most members of the rice XTH family exhibited organ- and growth stage-specific expression. This was confirmed by quantitative real-time reverse transcriptase-polymerase chain reaction analysis of representative OsXTH members. This revealed in more detail the temporally and spatially controlled expression profiles of individual OsXTH genes at particular sites in rice. Previous reports indicated that grasses have relatively greater xyloglucan endotransglucosylase activities, one of the two enzyme activities catalyzed by XTHs, than in equivalent tissues in dicotyledons. This observation, together with the tissue-specific and growth stage-dependent expression of a large rice XTH gene family, suggests that xyloglucan metabolism plays a more central role in monocotyledon cell wall restructuring than has been reported previously.
The most widely adopted models of the structure of primary cell walls in dicotyledonous plants view the hemicellulose xyloglucan as a structurally important glycan (Carpita and Gibeaut, 1993), cross-linking and tethering cellulose microfibrils, and thereby forming the basic load-bearing framework of these so-called type I walls (Carpita, 1996). Other models place less emphasis on xyloglucan cross-links but still envisage xyloglucan as interacting closely with the microfibrils (for review, see Cosgrove, 2000). Xyloglucans are found in type I cell walls in various tissues at different developmental stages, including the cell plates formed in dividing cells (Moore and Staehelin, 1988), the primary walls in growing cell, and fully differentiated secondary walls. Given its proposed important structural role, there is considerable interest in understanding the biochemical basis of xyloglucan synthesis, integration into the wall and subsequent modification: all processes that are considered to be a fundamental part of cell growth and differentiation.
The xyloglucan endotransglucosylase/hydrolases (XTHs) are a family of enzymes that specifically use xyloglucan as a substrate and that catalyze xyloglucan endotransglucosylase (XET) and/or xyloglucan endohydrolase activities. Thus, XTHs are thought to play an important role in the construction and restructuring of xyloglucan cross-links, although this has not yet been demonstrated. XTHs typically are encoded by large multigene families in dicotyledons (for review, see Nishitani, 1997; Rose et al., 2002); for example, in Arabidopsis, 33 open reading frames (ORFs) potentially encoding XTH proteins have been identified from the genome sequence database (Yokoyama and Nishitani, 2001b). Expression analysis of these genes has revealed that most of the family members exhibit distinct expression patterns in terms of tissue specificity and that they respond differently to hormonal signals (Xu et al., 1996; Akamatsu et al., 1999; Yokoyama and Nishitani, 2001a, 2001b; Nakamura et al., 2003). The ubiquitous occurrence of xyloglucans in various cell types, and the cell typespecific expression profiles of the XTH genes (Yokoyama and Nishitani, 2001b; Rose et al., 2002), together with recent studies of XET action in vivo (Vissenberg et al., 2000, 2001; Bourquin et al., 2002), indicate that XTHs are involved in a wide range of physiological processes. This is reflected in the large size of the Arabidopsis XTH (AtXTH) family, where each gene appears to play a particular role in modulating wall architecture in a temporally and spatially specific manner.
The primary cell walls from commelinoid monocotyledons, which include cereals such as rice (Oryza sativa), have a number of major structural and compositional differences from those of dicotyledonous species and are termed type II walls. Most studies report that type II walls have relatively little xyloglucan, and the predominant glycan that cross-links the cellulose microfibrils is instead glucuronoarabinoxylan (GAX; Carpita and Gibeaut, 1993). For example, xyloglucan and GAX account for approximately 20% and 5%, respectively, of walls from suspensioncultured cells from dicotyledons, whereas in cultured maize (Zea mays) cells, they account for approximately 5% and 40%, respectively, of the wall (Rose et al., 2000). The walls of cereals and grasses also contain (1,3)(1,4)-β-d-glucan (mixed-linkage glucan), which is considered to act as another cross-linking glycan. Moreover, the relatively small amounts of xyloglucans in type II walls are structurally quite different from those found in type I walls in terms of their size and branching patterns and are typically described as not being involved in cross-linking cellulose microfibrils (Carpita, 1996). Kato et al. (1982) showed that rice xyloglucans consisted of glucan backbones with single xylosyl residues without further substitution, the frequency of which is much less than that in dicots. Thus, rice contains a different type of xyloglucan as a minor component. Given the quantitative and qualitative differences in xyloglucan and its very different structural roles in the two distinct types of cell walls in flowering plants, it would be logical to predict that the XTH gene families of the two classes also would have evolved quite differently and that XTHs would be less numerous, diverse, and abundant in plants with type II walls.
The genome sequence of rice subsp. Japonica cv Nipponbare has been published recently (Goff et al., 2002) by the International Rice Genome Sequencing Project. The availability of this resource, together with the Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000), provides the first opportunity to undertake comparative phylogenetic analyses (Sasaki and Burr, 2000; Buell, 2003) of the whole complement of orthologs and paralogs of a given gene family in a dicotyledon and a commelinoid monocotyledon. Analysis of the draft sequences of the rice genome has revealed a large rice XTH (OsXTH) gene family with 29 members, a number that, contrary to expectations, is similar to that of the AtXTH gene family. This surprising finding raises interesting questions about the biological importance of XTHs in species with type II walls. To investigate the functions of the rice XTHs further, as well as their evolutionary significance, we have classified the 29 OsXTH genes on the basis of a systematic nomenclature and characterized the expression patterns of the whole complement of this gene family using DNA array expression profiling. The potential roles of these proteins and their substrate xyloglucans in plants with differing wall types are discussed.
RESULTS AND DISCUSSION
Identification of 29 Rice XTH Genes
The presence of XTH genes in the Poaceae was noted as early as 1993, when the first cloning of wheat XTH cDNA (originally termed TaEXT1) was reported (Okazawa et al., 1993). This was followed by the isolation and characterization of a maize XTH gene, the expression of which is up-regulated under flooding conditions (Saab and Sachs, 1996). More recently, four rice XTH genes were cloned and two of them implicated in contributing to internodal stem growth (Uozu et al., 2000). However, it was not until 2002 that an outline of the rice XTH gene family was suggested on the basis of the partially completed database of the rice genome sequence. Now that the draft sequence of the rice genome is almost complete, many putative genes encoding XTHs have been deposited through automated annotation processes and released from Web sites, as represented by the Rice Genome Program (Rice GAAS; http://rgp.dna.affrc.go.jp/giot/INE.html) and The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/e2k1/osa1; Table I).
Table I.
Identification of XTH genes in the rice genome
| Systematic Gene Name | Previous Gene Name | RiceGAASa | GeneBankb | Chromosomec |
|---|---|---|---|---|
| OsXTH1 | OJ2027_B02.20 | AP003899 | 7 | |
| OsXTH2 | OsXTR1 | OSJNBa0007D07.03 | AC136481 | 11 |
| OsXTH3 | OsXTR4 | OSJNBb0015D13.12 | AL606650 | 4 |
| OsXTH4 | OJ1217_D10.02 | AP003907 | 8 | |
| OsXTH5 | P0031C02.06 | AP004657 | 8 | |
| OsXTH6 | OSJNBa0041A02.39 | AL606638 | 4 | |
| OsXTH7 | OSJNBb0015N08.21 | AL662996 | 4 | |
| OsXTH8 | P0682A06.10 | AP004705 | 8 | |
| OsXTH9 | OSJNBa0041A02.36 | AL606638 | 4 | |
| OsXTH10 | P0028E05.21 | AP005445 | 6 | |
| OsXTH11 | OsXTR2 | P0028E05.28 | AP005445 | 6 |
| OsXTH12 | P0028E05.24 | AP005445 | 6 | |
| OsXTH13 | P0669G10.02 | AP005398 | 2 | |
| OsXTH14 | P0669G10.04 | AP005398 | 2 | |
| OsXTH15 | OSJNBa0012F14.38 | AP004784 | 6 | |
| OsXTH16 | OSJNBb0015N08.20 | AL662996 | 4 | |
| OsXTH17 | P0682A06.21 | AP004705 | 8 | |
| OsXTH18 | P0028E05.26 | AP005445 | 6 | |
| OsXTH19 | OJ1384D03.12 | AC113930 | 3 | |
| OsXTH20 | OSJNBa0001O14.09 | AC025783 | 10 | |
| OsXTH21 | OJ1624_G09.09 | AP003839 | 7 | |
| OsXTH22 | OJ1136_C04.15 | AP004026 | 2 | |
| OsXTH23 | OsXTR3 | B1053A04.21 | AP005859 | 2 |
| OsXTH24 | OSJNBb0060O08.26 | AC120506 | 3 | |
| OsXTH25 | OSJNBa0027L23.23 | AC018929 | 10 | |
| OsXTH26 | P0576F08.35 | AP004886 | 2 | |
| OsXTH27 | OSJNBa0023I19.12 | AP079037 | 10 | |
| OsXTH28 | OSJNBa0060E18.16 | AC118981 | 3 | |
| OsXTH29 | P0705B06.23 | AP005430 | 9 |
Gene no. assigned by RiceGAAS (http://RiceGAAS.dna.affrc.go.jp, February 6, 2003)
GenBank accession nos. of the Genomic DNA
Chromosome no
A list of putative XTHs was compiled by searching these databases for ORFs with characteristic sequences that are highly conserved among XTHs. Some ORFs were rejected because they contained uncharacteristic amino acid sequences located in the predicted C-terminal regions or lacked the signature DEIDFEFLG motif (see Fig. 1) that includes the residues responsible for the catalytic activity and is conserved among all the XTHs thus far characterized (Yokoyama and Nishitani, 2001b; Rose et al., 2002). It was apparent that in several cases, the gene structures annotated by the Rice GAAS do not match those annotated by TIGR (http://www.tigr.org/tigrscripts/tgi/T_index.cgi?species=rice). In these cases, the coding regions of these predicted genes were manually reinterpreted based both on the conserved structural features of XTHs and on the sequence database of full-length cDNAs released from the Knowledge-Based Oryza Molecular Biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/). After full analysis and re-annotation, 29 unique ORFs encoding putative rice XTH (OsXTH) genes were identified (Fig. 1). Four of the 29 genes had already been reported as OsXTR1, OsXTR2, OsXTR3, and OsXTR4 (Uozu et al., 2000), and these were renamed with permission from the original authors (Uozu et al., 2000) according to the recently proposed unified nomenclature system for XTH genes (Yokoyama and Nishitani. 2001b; Rose et al., 2002). The finalized nomenclature of these four genes, together with the rest of family, is listed in Table I and at http://labs.plantbio.cornell.edu/XTH.
Figure 1.
Alignment of the amino acid sequences of 29 OsXTH proteins, constructed using the ClustalW program and drawn with the Boxshade program. Identical and similar amino acid residues are shown on black and gray backgrounds, respectively. The DEIDFEFLG motifs are indicated by letters on top of the alignment. Signal peptide sequences, as predicted by the PSORT program, are represented in lowercase.
PSORT analysis (http://psort.nibb.ac.jp/) predicted a signal peptide for entry into the secretory pathway for each of the 29 putative OsXTH proteins. All the OsXTHs possess the diagnostic amino acid sequence motif DEIDFEFLG, which is conserved not only in XTH proteins (Nishitani, 1997) but also in the family 16 glycoside hydrolases, a class of hydrolases that includes β-glucanases from members of the Bacillus genus (Henrissat et al., 2001). In 26 of the OsXTH proteins, one or more N-linked glycosylation sites were identified adjacent to the C-terminal side of the DEIDFEFLG motif, whereas no glycosylation site was found in OsXTH19, -20, or -21 (Fig. 1).
Phylogenetic Relationships between OsXTHs and AtXTHs
To investigate the evolutionary relationships of the XTH family of genes in rice and Arabidopsis, a phylogenetic analysis was performed using full-length protein sequences (Fig. 2). It was shown previously that AtXTH members could be classified into three groups and this classification has been applied to a broad range of plant species (Nishitani, 1997; Campbell and Braam, 1999b; Yokoyama and Nishitani, 2001b; Rose et al., 2002). However, upon superimposing the dendrogram of OsXTHs over that of AtXTHs, the divergence between Groups I and II of the AtXTH was no longer apparent. However, the third group was clearly distinct from Groups I and II, indicating that the XTH genes could be divided appropriately into two major subfamilies: Group I/II and Group III. Given the two-subfamily classification for the XTH gene family, the Group I/II subfamily contained 18 OsXTH genes and 26 AtXTH genes, whereas the Group III subfamily consisted of 11 OsXTH genes and seven AtXTH genes. In dicotyledons, members of Group III have been shown to catalyze xyloglucan hydrolysis (xyloglucan endohydrolase activity) rather than xyloglucan transglucosylation (XET activity; Farkas et al., 1992; Fanutti et al., 1993; Tabuchi et al., 2001). However, it has not been demonstrated strictly that all members of this group exhibit this characteristic. Apparently, rice contains a greater number of Group III members than Arabidopsis, although the functional significance of this is unclear.
Figure 2.
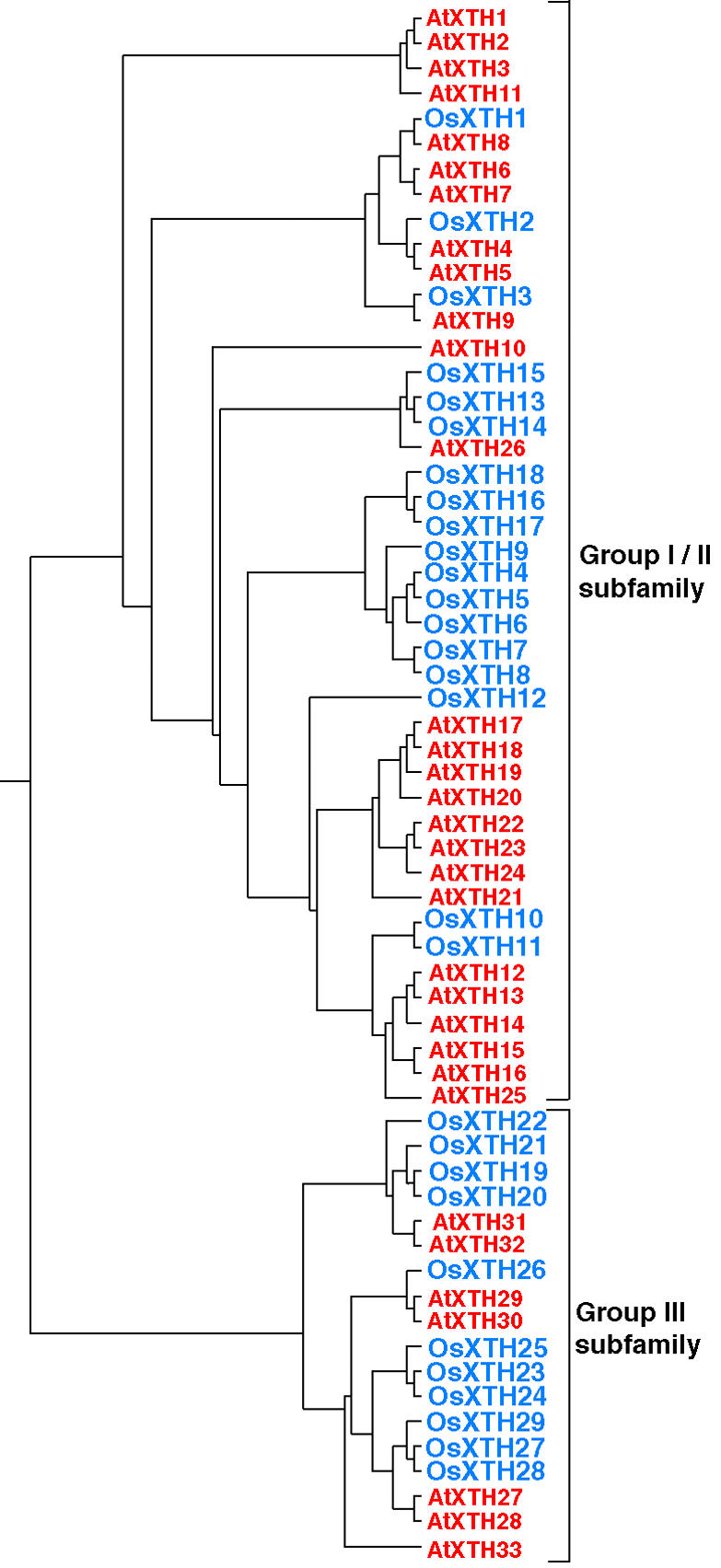
Dendrogram generated using ClustalW and TreeViewPPC software, based on the predicted amino acid sequences of AtXTHs and OsXTHs, which are shown in red and blue, respectively. The XTH members are classified into two subfamilies based on the classification for Arabidopsis XTH genes in which the AtXTH members are classified into three groups (Yokoyama and Nishitani, 2001b).
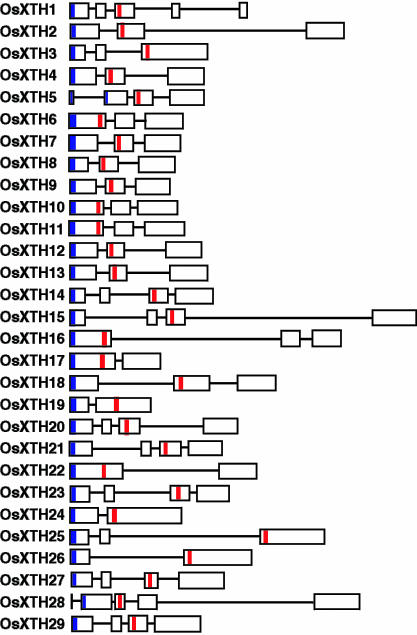
The structures of individual OsXTH genes were determined based on their genomic sequences. With few exceptions in Arabidopsis XTHs, gene structure, such as exon organization, is well conserved within each group. For example, one feature common to AtXTH Group II genes is that the well-conserved DEIDFEFLG motif is located in the second exon (Yokoyama and Nishitani, 2000, 2001b; Rose et al., 2002). In rice XTHs, on the other hand, the number of exons varies from two to five and is independent of the subfamily (Fig. 3). Furthermore, unlike Arabidopsis, the DEIDFEFLG motif is not confined to the second exon but is found in either the first, second, or third exons. Of the 14 OsXTH genes that consist of three exons, four genes encode the motif in the first exon, and there are eight genes in the second and three genes in the third (Fig. 3).
Figure 3.
A schematic diagram of the OsXTH gene structures. White boxes, exons; lines, introns; Blue boxes, Putative signal peptide sequence; red boxes, DEIDFEFLG motifs that correspond to the catalytic sites of the proteins.
Expression profiles of OsXTH Genes
Because members of the OsXTH gene family share similar sequences in their coding regions, a sequencespecific detection procedure is required for the analysis of their expression. To satisfy this requirement, an oligonucleotide microarray was used that allows both specific measurements and a comprehensive survey of the mRNA levels of the whole complement of OsXTHs. For this purpose, and based on the 3′ non-coding regions of OsXTH genes, aminomodified 70-nucleotide probes were designed that were specific for individual OsXTH genes (Table II). To generate OsXTH-specific microarray slides, these amino-modified 70-nucleotide probes were spotted and immobilized on the surface of TaKaRa-Hubble glass slides (TaKaRa Biosciences Co., Ltd., Ohtsu, Japan) that had been coated with activated polyesters capable of covalently linking amino-modified oligonucleotides with high efficiency.
Table II.
Gene-specific probes used for oligoDNA microarray
| Gene Name | probe Sequence (5′ to 3′) |
|---|---|
| OsXTH1 | GCATGGATGTCAGAGAAAAAAATAATGTTTCTCTCCTGCCACAGATCTGGGTGATGAAGGTGAGGATAGG |
| OsXTH2 | GATCTCATCGAGATCTGTGATCGATGCTTGCTGGCCGTCCGATGGATGGATGGATGGATGGGGATGAGTA |
| OsXTH3 | AACTGCTGGTTTCTTTCTTGGTTAGCAAATCAAATGGCAGGTAGTTTATCTCAGATGAACTTAGGTGATG |
| OsXTH4 | AAGTAATTCAATTAGCAGATAGAAAGGGTGAGAGGTGTATTGGATGATTCGGTTCATTCTTTCCAAGTAT |
| OsXTH5 | AAGTATTTCAGTTAGCTGATAGAAAGGGTAGCTGAGTTTTTTTTTTCTTTTGCACATTTGTTTGAGGAAA |
| OsXTH6 | GATAGAGGTGGCTCGGTATGCATGTGAGAAGTGCGTGAAATGTACCGAATCAATATTCCTTTTTTGAAGA |
| OsXTH7 | GAAGAAGATCGAATTGATCGAGGGGATCGATCGATTGCATGCTGATTCATCATTCGTTATTCTTGTACAT |
| OsXTH8 | CTAGTTATAGCTAGAGAGCGTGTTTAGTTTAGTCCAAAATTTCTTAATTAATTCGTGTGTAATTTCTCTC |
| OsXTH9 | AGTGGAGTGGAGACGCGCCAAAGCCTCGGTGCACTTTGATGAACGCGGACTCGATCGAACTCACGCAGGG |
| OsXTH10 | TAGTAAGAATATCCCATCGTCGTCGTCGTCGTCGTCGTCGATCGCCGGCGGAGCAAGATGGCCGGAGATG |
| OsXTH11 | GATCGACACGAGAAGATGATCATAACGTACGCTACGCATGCATGATCCGTCATCGATGAAAAAAATGATG |
| OsXTH12 | GGGGCACTTGAAACTATTTTATTGTAGAATTATTCTGTATCTATTGCTGTATGTGTTATTGTCGTGTAAA |
| OsXTH13 | GTGCTGAAGGATCGATATTGCAATTGCAAAGTTTTTGCTATATGTGCAATCATCTTTTGAGAGATGTTCC |
| OsXTH14 | TTTACCAGCTGCTATGAGTATGGATATTTTTTATGAGTTTTGAGATTTATTTGATGATTTTTTTTCTATT |
| OsXTH15 | TCATGCATGCGCATGTCTAGTTTCACTTGATTTTTCATGGCATGTTTGATTAATGGCTTCTTGGAAGAAG |
| OsXTH16 | CCGGAGCCGGCCATTGCCTCCGTGCTGTTCTAGCATTCACGATACTGTAACTTGATCTCTACTGTACTTC |
| OsXTH17 | TTTTGAACTCGATCGATTCAAATCCTCCTCCATTGATGAGTTCTTGGCAATGATTTGTAATTGCTTCTTG |
| OsXTH18 | GGCGTCGTACGCGACGTCGACTCCATGATGATTCCATCCGTCGTCCAGCGATCTGTAGGCTCATACTTGT |
| OsXTH19 | TGGTTGAGCCATGCATCTTCAGTTTCACATCGTTTCATGTGTTGTGTGCACCACGTAGTACGAGCACCAA |
| OsXTH20 | TGAACAGCGTCAAGATTATTGCTCATTGGCTCATTGCTTCGTTGGATTCATGTGAAGTGTGATCGAATTA |
| OsXTH21 | CGAGTGCCGTTGCCGTGCTGATTCGTGTGTGTTGTGTTCTTGTTTGGATTTTGGGAAAGGTAATGTGGGT |
| OsXTH22 | CTAACTACTGCTTAATTATCTAATACGCCGCCATGTATACTCCATCTAATACGCTCCTCCGATCCTCAAT |
| OsXTH23 | AGTGTCAACTTGGGAAGCTAATGGCATATGATGGTTACTATTACACTACTAGATTCTATTAATTTTGCGG |
| OsXTH24 | TCTTTGATTAGAATATTCTATTCTATGATGAGGCTCTAGCTCCATCATAATTCTTTGATTAATTTTGCTT |
| OsXTH25 | TTTATTTACTAGTACTGTGTAATCTCAATCATGCATTATTCCACCAGTTGATCCTGCTTCTAGATGATCG |
| OsXTH26 | CGCAGCACATAATCATATTTGTTTCATCATTTCATCGTCGCGCCTTCGAGTCATGGGATGATCGAAACGG |
| OsXTH27 | AGCATTCTGATCATCACAATTGGCTCAGTGTGTGCTCACCTCCCTTAATTTTGTGTGGCTTATGTGATGA |
| OsXTH28 | CCGATTCTGTGATCCGCACAGCGCCGCTCGCGATGGTGGCGCGCGTGCTCGACTGTACTATCGTGCCGTG |
| OsXTH29 | TACAGTAAGCATCAGCAGTGGGAACGCGTCGAGGCGCCGCTCGCCGCTCGCCGCCGTGGCAGACAGGCGT |
| OsActin | TTCTTCGGACCCAAGAATGCTAAGCCAAGAGGAGCTGTTATCGCCGTCCTCCTGCTTGTTCTCTCTTTTT |
The oligo DNA microarray was used to examine the expression levels of all OsXTH mRNAs in seedlings that had been grown for 3, 7, or 14 d. The endosperm was first removed from the seedling leaving coleoptiles, leaves, and roots. These growth stages were chosen because vigorous cell expansion occurred during these periods, both in the shoots and roots. At least three microarray hybridizations were performed for each assay point using Cy3/Cy5-labeled cDNA that had been independently reverse transcribed from equal quantities of mRNAs from individually harvested rice plants. The expression levels of individual OsXTH genes were determined from the Cy3/Cy5 fluorescence intensity ratios in the microarray image, which were normalized using a rice actin gene (accession no. AU091532) as an internal standard. Transcription levels of the actin gene in the same RNA samples used for the microarray analysis were determined by quantitative real-time reverse transcriptase (RT)-PCR analysis.
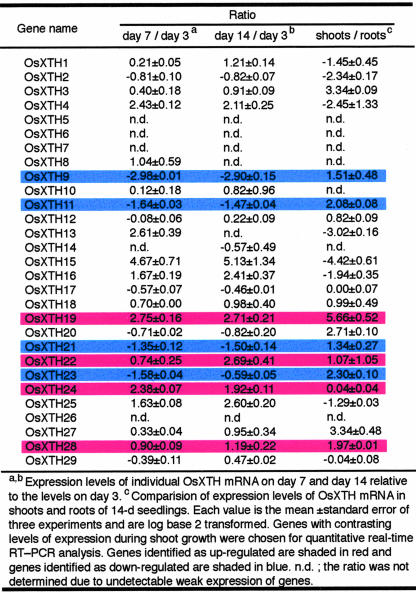
The microarray hybridization analysis revealed that 25 members of the 29 OsXTH gene family members found in the rice genome were expressed in the rice plants used in this study; 21 genes were transcribed at relatively high levels, and four genes (OsXTH8, -10, -13, and -14) were expressed at lower levels (Table III). These results indicate that the 25 OsXTH ORFs are actually transcribed, and the corresponding genes are functional. Expression of the remaining family members, OsXTH5, -6, -7, and -26, was not detected under the conditions used (Table III). At present, it is not known whether or not these four genes are functional because they might be expressed in minor tissues that were not examined during specific developmental stages or in response to biotic or abiotic stimuli that were not used in the present study.
Table III.
Expression patterns of the OsXTH
Table III also indicates that the expression of most members of the OsXTH gene family exhibit growth stage-dependent expression patterns; the expression of OsXTH9, -11, -21, and -23 decreased as growth proceeded, whereas that of OsXTH1, -4, -15, -16, -19, -22, -24, -25, and -28 increased during the same period.
To examine the organ-specific expression profiles of these genes, a comparison was made of the expression levels in shoots, which are composed of coleoptiles and leaves, and in roots in the 14-d-old seedlings with the endosperms removed (Table III). Although seven OsXTH genes (OsXTH1, -2, -4, -13, -15, -16, and -25) were expressed preferentially in roots, there was with little or no expression in the shoots, which suggested that their expression was root specific. In contrast, seven OsXTH genes (OsXTH3, -11, -19, -21, -23, -27, and -28) were predominantly expressed in the shoots. OsXTH24 was expressed equally in both tissues.
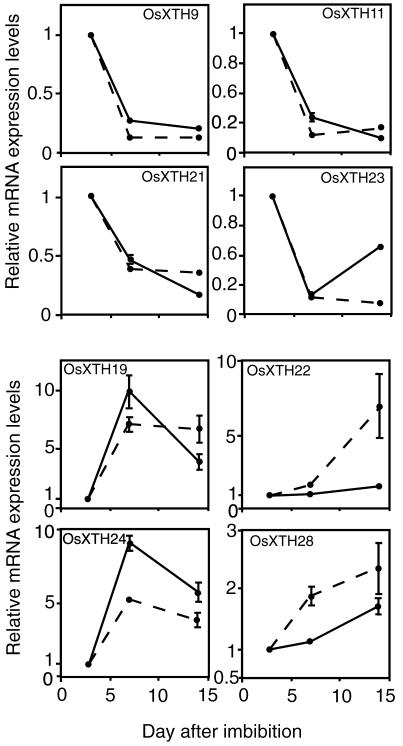
To more precisely quantify transcript levels of the OsXTH genes whose expression patterns were found to change during shoot growth, the expression profiles of eight representative genes (OsXTH9, -11, -19, -21, -22, -23, -24, and -28) were analyzed during shoot growth, from d 3 to 14, by quantitative real-time RT-PCR (Fig. 4). These genes were chosen because their expression levels were found to decline (OsXTH9, -11, -21, and -23) or rise (OsXTH19, -22, -24, and -28) during shoot growth (Table III). The results obtained using the RT-PCR experiments were consistent with those obtained with the microarray analysis, as seen by comparing the solid versus the broken lines in Figure 4. The only differences in the magnitudes and kinetics of the changes in expression levels were seen with OsXTH22 and -23. Thus, the results validate the usefulness of the oligonucleotide DNA microarray procedure.
Figure 4.
Quantitative analysis by real time RT-PCT of transcript abundance of eight representative OsXTH genes expressed in 3-, 7-, and 14-d-old seedlings of Arabidopsis. Ratios of transcript levels in 7-and 14-d-old seedlings to those in 3-d-old seedling are shown. To compare the data obtained by the real-time RT-PCR analysis with those obtained by the microarray analysis, the data shown in Table III are superimposed as broken lines. Error bars = se based on three replicates of the same sample.
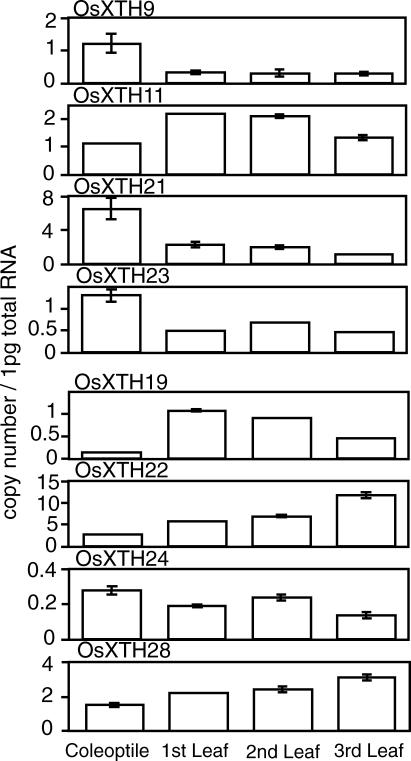
Real-time RT-PCR was also used to determine the transcript levels of the eight genes in coleoptiles and the first to third leaves at particular growth stages when the respective organs were at their early mature stages (Fig. 5). Three OsXTH genes (OsXTH9, -21, and -23), exhibited higher expression in the coleoptiles than in the leaves. The expressions of these three genes in the whole seedlings decreased from d 3 to 14 (Fig. 4). It should be noted that the d 3 shoots were composed chiefly of coleoptiles and small juvenile leaves within the coleoptiles and that as the growth stage proceeded from d 3 to 14, leaves successively developed, and the coleoptiles senesced. Thus, the decline of the expression levels of the three genes in the whole seedling from d 3 to 14 might reflect changes in organ weights occurring during shoot development, i.e. shrinkage of the coleoptiles by aging and the massive leaf development. On the other hand, three other XTH genes (OsXTH19, -22, and -28) whose transcript levels in whole shoots were found to increase during seedling growth showed higher expression levels in the leaves than in the coleoptiles. Again, this transition of expression patterns might be attributed to the development of leaves during the growth stage (Fig. 5).
Figure 5.
Expression levels of the eight OsXTH genes in coleoptiles and leaves. Coleoptiles and first to third leaves were excised separately from 3-to 14-d-old seedlings and subjected to RNA extraction. Transcript levels were determined by quantitative real-time RT–PCR and are shown as the number of RNA molecules in 1 pg of total RNA. Error bars = se based on three replicates of the same sample.
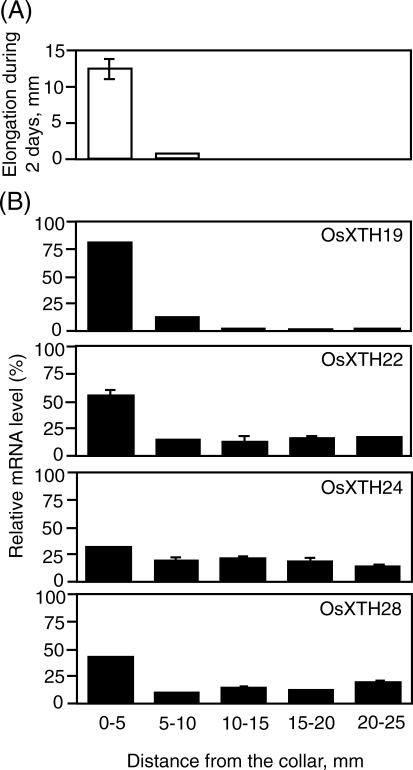
To more precisely examine the correlation between cell expansion and transcript abundance of the OsXTH genes whose expression was up-regulated during shoot development (OsXTH19, -22, -24, and -28), more detailed expression analysis was performed in the third leaves (Fig. 6). Figure 6 shows the spatial pattern of blade elongation (Fig. 6A) and transcript abundance of the four OsXTH genes along the 25-mm-long blade of the third internodes (Fig. 6B). The elongation zone was restricted to the basal 5-mm region above the collar and little or no elongation was observed in the upper regions (Fig. 6A). Of the four genes, OsXTH22, -24, and -28 were predominantly expressed in the elongation regions (0–10 mm above the collar), whereas OsXTH24 was expressed equally along the blade. OsXTH19 was expressed almost exclusively in the basal 10-mm region and showed the greatest correlation with the blade elongation rate, implying a role for OsXTH19 in leaf cell expansion.
Figure 6.
Expression profiles of four OsXTH genes along the third leaf in 14-d-old plants. A, Elongation of each region of the leaves during a 2-d period. B, Transcript levels of the four genes in individual regions along the third leaf as measured by quantitative real-time RT-PCR analysis. The relative abundance of individual mRNAs in each region is shown as percentages of the total copy number in the whole leaf. Error bars = se based on three replicates of the same sample.
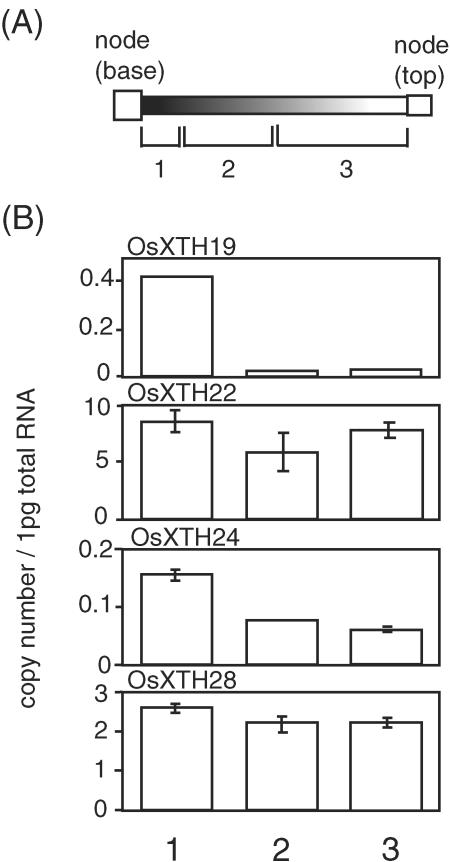
Finally, the expression of the same four OsXTH genes was examined in the internodes of mature 12-week-old rice plants. Figure 7 shows the abundance of the gene transcripts in the three regions of the internodes: the dividing/elongating zone at the basal region, the premature nonelongating zone in the middle region, and the mature nonelongation zone in the upper region. Interestingly, OsXTH 19 was specifically expressed in the dividing/elongating zone, suggesting that it functions in cell expansion in both leaves and internodes of rice plants.
Figure 7.
Expression profiles of four OsXTH genes along the internodes in 12-week-old plants. A, Schematic description of the three zones, defined as: 1, the dividing/elongating zone with intercalary meristem; 2, premature nonelongating zone; and 3, mature nonelongating zone (Uozu et al., 2000). B, Transcript levels of the four OsXTH genes in regions of the internodes as determined by quantitative real-time RT-PCR. The transcript levels are shown as the number of RNA molecules in 1 pg of total RNA from each sample. Error bars = se based on three replicates of the same sample.
In conclusion, the combined expression profiles of the OsXTH genes, particularly those expressed in the shoots, reveal that they are expressed at specific sites and that each exhibits a unique “expression fingerprint,” implying involvement in divergent and specific processes. What, then, are their biological functions?
Biological significance of OsXTHs in Xyloglucan-Poor Rice Cell Walls
In Arabidopsis, 33 putative AtXTH genes have been identified and classified on the basis of their genome structures and occurrence in the expressed sequence tag (EST) database (Yokoyama and Nishitani, 2001b). These genes are thought to encode proteins that are responsible for either the molecular grafting or disassembly of xyloglucan molecules in the walls of various cell types. The ubiquitous nature of XTH expression and the extensive size of the gene family probably reflect the diverse roles of XTHs and the fact that their specific substrate is the principal hemicellulose in type I walls. In contrast, relatively far less xyloglucan is present in the primary cell walls of commelinoid monocotyledons, such as is rice, and its structure is different from that of xyloglucan in type I walls (Kato et al., 1982; Carpita and Gibeaut, 1993). As a consequence, current models of type II wall architecture do not attribute a structurally important role to xyloglucan but, rather, suggest that GAX and perhaps mixed-linkage glucan cross-link the cellulose microfibrils, thereby forming the key load bearing framework. Therefore, it is surprising that not only is the number of OsXTH family members comparable with that of the AtXTH family, but that most members of the OsXTH family are expressed and in a pattern that is similar to that seen in Arabidopsis, with the majority showing increased mRNA expression levels during elongation growth but a few declining in abundance. Moreover, previous studies in rice (Uozu et al., 2000) and other monocotyledons such as barley (Hordeum vulgare; Smith et al., 1996) and maize (Pritchard et al., 1993; Wu et al., 1994; Palmer and Davies, 1996) also have seen correlations between tissue growth rate and XTH mRNA abundance and/or XET activity.
However, perhaps an even greater paradox is that two surveys of a phylogenetically broad range of plant species indicate that total extractable XET enzyme activity in both growing aerial (Fry et al., 1992) and root (Wu et al., 1994) tissues is considerably greater in grasses than in dicotyledons, based on either a fresh weight or specific activity basis. If xyloglucan has only a minor structural role in type II walls, as is asserted in current models, why are the XTH gene family complexity, mRNA expression levels, and enzyme activity in monocotyledons comparable with, or even more pronounced, than in species with type I walls?
One likely explanation is that the xyloglucan in type II walls has a more important structural function than is generally described and that although quantitatively less abundant on a relative basis because of the presence of substantial amounts of GAX, xyloglucan polymers make a significant contribution to wall properties, whether as inter-microfibril cross-links or in some other conformation. In this way, this population of xyloglucans might be targeted for modification during cell wall restructuring in conjunction with other polymers such as GAX. Consistent with this idea, Inouhe et al. (1984) have shown that Mr of xyloglucan in coleoptile sections of oat (Avena sativa), a Poaceae, substantially decreases when auxin induces their elongation growth, a typical auxin action that was demonstrated in Vigna angularis, a dicotyledonous plant (Nishitani and Masuda, 1981). It is not clear why rice has a proportionally greater number of XTH genes in the Group III subfamily, a group that has been associated with xyloglucan hydrolysis (Rose et al., 2002). The functional basis of the subfamily grouping is still unknown and will remain so until substantial biochemical data describing the cognate enzyme activities have been generated. Furthermore, the higher levels of XET activity in the Poaceae, in combination with a far less abundant xyloglucan substrate, might argue for a more, rather than less, important role, all other things being equal. A caveat to this observation is that the enzyme activities (Fry et al., 1992; Wu et al., 1994) were not assayed using the native xyloglucan substrates, although the differences are still striking.
An alternative possibility is that xyloglucan does play a fairly insignificant structural role in type II walls and that the abundance of XTH genes and related mRNA expression and enzyme activities reflects xyloglucan modification throughout the plant that results in subtle and not structurally critical wall reorganization. This seems unlikely because in this case one might predict that the OsXTH and AtXTH gene families would exhibit substantial differences in gene number, organization, and expression.
A third explanation is that in some circumstances, XTHs may be involved in the modification of hemicelluloses other than xyloglucan. As stated previously, rice cell walls contain large amounts of GAX and (1–3),(1–4)-β-d-glucan (Carpita and Gibeaut, 1993; Carpita, 1996), and whereas all XTHs from dicotyledons tested to date specifically use xyloglucan as a substrate (e.g. Fry et al., 1992; Nishitani and Tominaga, 1992), it is conceivable that XTHs from rice and other monocotyledons may act on other polymers, such as (1–3),(1–4)-β-d-glucans. To our knowledge, no studies have been reported that have addressed the substrate specificity of XTH proteins from a commelinoid monocotyledonous species. A three-dimensional structure, although imminent (Johansson et al., 2003), has yet to be published for any XTH, limiting predictions of the relative orientation of different polysaccharides relative to the proposed XTH catalytic site. The DEIDFEFLG motif that is critical for cleavage of the β-1–4 linkages in the xyloglucan backbone (Campbell and Braam, 1999a) is conserved among most XTHs but shows some divergence in specific predicted XTHs from both monocotyledons and dicotyledons (Fig. 1). Variation in amino acid residues at and around the catalytic site is likely to cause a significant change in the mode of enzyme action and potentially substrate specificity. To address this issue, comparative biochemical analysis of individual XTH proteins from species with type I and type II walls will be necessary. Such an approach, in combination with cell wall analysis, might offer an opportunity to explore new aspects of XTH function in plants, particularly in the context of different cell wall types.
MATERIALS AND METHODS
Database Search for Sequence Analyses
Sequence information for genes, proteins and cDNAs was retrieved by searching public databases with a keyword search or BLAST algorithm at the Rice Genome Research Program (http://rgp.dna.affrc.go.jp), the TIGR Rice Genome Project (http://www.tigr.org/tdb/e2k1/osa1/), and the Knowledge-Based Oryza Molecular Biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/). To ensure accuracy in the annotations of the rice (Oryza sativa) XTH genes, they were reexamined by three procedures: multiple sequence alignments of predicted amino acid sequences, comparison of the genomic coding sequences and corresponding full-length cDNA sequences, and verification of the correctness of the ORF and intron organization on the basis of the conserved structural features of the XTH genes thus far identified. The protein localization site was predicted using the PSORT program (http://psort.ims.u-tokyo.ac.jp). Multiple sequence alignments were constructed using ClustalW (http://spiral.genes.nig.ac.jp/homology/) and were displayed with the software available at the Boxshade Web site (http://www.ch.embnet.org/). The dendrogram was generated using ClustalW and TreeViewPPC software, based on the predicted OsXTH and AtXTH amino acid sequences.
Preparation of Oligonucleotide Microarray for OsXTH Genes
Oligonucleotide sequences that were specific for individual OsXTH genes were designed based on the 3′-untranslated region (UTR) of each gene, from sequences in the EST, full-length cDNA, and genome databases. The putative 3′-UTR region of any genes that were not found in an EST or full-length cDNA database were predicted based solely on the genomic information. The 70-nucleotide oligonucleotides from the 3′-UTR regions of individual OsXTH genes were designed such that they did not share more than 60% identity with equivalent regions of other OsXTH genes. In cases where such specific sequence was not found in the 3′-UTR, sequences within the 5′-UTR were selected so that the degree of homology was minimized. The 70-nucleotide oligonucleotides were synthesized with amino-modified 5′ termini, then spotted and immobilized onto TaKaRa-Hubble slides using an OmniGridder printer (GeneMachines, San Carlos, CA) by Hokkaido System Science Co., Ltd. (Sapporo, Japan) to generate OsXTH microarray slides. Negative charges inherent in the polyester surface of the microarray slide prevent nonspecific adsorption of negatively charged nucleic acids. This results in reduction of the background signal because of nonspecific binding of Cy3/Cy5 dye-labeled compounds, thereby increasing the signal-to-noise ratio. A broader dynamic range was achieved compared with conventional cross-linking procedures using polycarbodiimide-mediated immobilization.
Plant Material
Rice subsp. japonica cv Nipponbare seeds were germinated under water and water cultured for 3, 7, or 14 d in a growth chamber at 28°C with a day period of 14 h under fluorescent lamps at a photon flux density of 500 μmol m-2 s-1. The 3-, 7-, and 14-d-old seedlings were harvested, frozen in liquid nitrogen, and subjected to total RNA extraction. To investigate organspecific gene expression, the 14-d-old seedlings were divided into shoots and roots, which were separately subjected to RNA extraction. To analyze the correlation between leaf development and gene expression, the third leaves in the 14-d-old seedlings were selected for uniformity of blade length (25 mm) and divided into 5-mm regions and either excised and immediately frozen in liquid nitrogen for subsequent RNA extraction or marked with ink and allowed to grow under the same conditions for an additional 2 d, followed by measurements of the lengths, to assess growth rate. For analysis of the internodes, the 14-d-old seedlings were transferred to a pot with soil and grown under the same light and temperature conditions for an additional 10 weeks. The second or third internodes of the 12-week-old plants were divided into dividing, elongating, and nongrowing zones according to the procedure described by Uozu et al. (2000) and then frozen before RNA extraction. Total RNA was extracted according to the method of Chomczynski and Sacchi (1987).
Preparation of Fluorescent Probes and Microarray Hybridization
Cy3- and Cy5-labeled cDNA probes were prepared using the CyScribe Post-Labeling Kit (PRN5660, Amersham Biosciences Corp., Piscataway, NJ). In brief, 50 μg of total RNA from each sample was used as the template for reverse transcription, which was performed using oligo(dT) as the primer and a nucleotide mix containing amino-allyl dUTP. Thus, the first strand cDNAs synthesized were purified by alkali treatment followed by column purification using the CyScribe GFX purification kit and reacted with monofunctional Cy3/Cy5 reactive dye in the presence of sodium bicarbonate (pH 9.0). The Cy3/Cy5-labeled cDNAs were purified using AutoSeq G-50 columns (Amersham Biosciences), dried, and dissolved in 6.5 μL of water.
Hybridization and washing of the DNA microarray were performed as recommended by the manufacturer (Amersham Biosciences) with modifications as follows. The Cy3/Cy5-labeled cDNA solutions (6.5 μL) were mixed with 1 μL of salmon sperm DNA solution (0.1 μg μL-1), 7.5 L of hybridization buffer, and 15 μL of 100% (w/v) formamide, followed by hybridization at 42°C overnight. After hybridization, the microarray slides were washed successively with a solution containing 2× SSC and 0.1% (w/v) SDS at room temperature for 10 min, three times with a solution containing 2× SSC and 0.1% (w/v) SDS at 55°C for 10 min, and finally three times with a solution containing 0.1× SSC and 0.1% (w/v) SDS at room temperature for 10 min. Immediately after the washes, the hybridized slides were dried by spinning in a centrifuge.
Image Acquisition and Analysis
The hybridized and washed microarray slides were scanned at 570 nm for Cy3 signals and at 660 nm for Cy5 signals using a laser fluorescence scanner (GenePix 4000B, Amersham Biosciences). The fluorescence intensity data were acquired with the photomultiplier gain set at the highest level, and the data were processed by GenePix Pro version 3.0 software (GenePix 4000B, Amersham Biosciences) to subtract the local background on the microarray slides. The Cy3 and Cy5 signal images were normalized using the signals for the actin gene as an internal standard. The expression level of the actin gene per amount of total RNA in the same sample was quantified by real-time RT-PCR using the primers 5′-ttcttcggacccaagaatgc-3′(forward) and 5′-aacaaaagagagaacaagcaggag-3′(reverse). Microarray hybridization was repeated at least three times for each experiment, and the mean values of three experiments are given with ses.
Quantitative Real-Time RT-PCR
The oligonucleotide primers used for the RT-PCR analysis were designed based on the nucleotide sequences in the 3′-UTR of the eight OsXTH genes (OsXTH9, -11, -19, -21, -22, -23, -24, and -28) according to the procedure described previously (Yokoyama and Nishitani, 2001b). The following sequences correspond to primers used for the RT-PCR analysis: OsXTH9, 5′-agtggagtggagacgcgc-3′ (forward) and 5′-cctgcgtgagttcgatcga-3′(reverse); OsXTH11, 5′-tgatcataacgtacgctacgca-3′(forward) and 5′-caaaacacattcattattcgcca-3′(reverse); OsXTH19, 5′-gccatgcatcttcagtttcaca-3′(forward) and 5′-aatagtagatacgtgttggtgctcgt-3′(reverse); OsXTH21, 5′-agtggattcacgagtgccg-3′(forward) and 5′-ttacctttcccaaaatccaaaca-3′(reverse); OsXTH22, 5′-tacgccgccatgtatactcca-3′(forward) and 5′-caaacgattaattgacaaacaggg-3′(reverse); OsXTH23, 5′-gcatatgatggttactattacactactagattctat-3′(forward) and 5′-ctagcac-ctactacctttacaaactatacataca-3′(reverse); OsXTH24, 5′-ggctctagctccatcataattctttg-3′(forward) and 5′-tcgtgaagaagcagcagtaacaa-3′(reverse); and OsXTH28, 5′-gccgctcgcgatggt-3′(forward) and 5′-ggaaactataatcacgtacgacatcac-3′(reverse). Quantitative one-step RT-PCR was performed using a SYBR Green RT-PCR Reagents kit in an ABI Prism TM 5700 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA) according to the protocol provided by the supplier.
Acknowledgments
We thank Professor Makoto Matsuoka (Nagoya University, Chikusa, Nagoya, Japan) for his invaluable discussions and helpful suggestions concerning the growth pattern of rice plants.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.035261.
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant-in-Aid for Scientific Research on Priority Areas no. 15031202 and Grant-in-Aid for Scientific Research [B] no. 15370016) and by the Program for “Development of Fundamental Technologies for Controlling the Process of Material Production of Plants” from the New Energy and Industrial Technology Development Organization (Japan).
References
- Akamatsu T, Hanzawa Y, Ohtake Y, Takahashi T, Nishitani K, Komeda Y (1999) Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis. Plant Physiol 121: 715-721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14: 3073-3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR (2003) Current status of the sequence of the rice genome and prospects for finishing the first monocot genome. Plant Physiol 130: 1585-1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Braam J (1999a) In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J 18: 371-382 [DOI] [PubMed] [Google Scholar]
- Campbell P, Braam J (1999b) Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 4: 361-366 [DOI] [PubMed] [Google Scholar]
- Carpita NC (1996) The structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445-476 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1-30 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156-159 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Expansive growth of plant cell walls. Plant Physiol Biochem 38: 109-124 [DOI] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid J (1993) Action of a pure xyloglucan endotransglycosylase (formerly called xyloglucan-specific endo-(1–4)-beta-d-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J 3: 691-700 [DOI] [PubMed] [Google Scholar]
- Farkas V, Sulova Z, Stratilova E, Hanna R, Maclachlan G (1992) Cleavage of xyloglucan by nasturtium seed xyloglucanase and transglycosylation to xyloglucan subunit oligosaccharides. Arch Biochem Biophys 298: 365-370 [DOI] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglycosylase, a new wall-loosing enzyme activity from plants. Biochem J 282: 821-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T-H, Presting D, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. Japonica). Science 296: 92-100 [DOI] [PubMed] [Google Scholar]
- Henrissat B, Coutinho PM, Davies GJ (2001) A census for carbohydrateactive enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55-72 [PubMed] [Google Scholar]
- Inouhe M, Yamamoto R, Masuda Y (1984) Auxin-induced changes in the molecular weight distribution of cell wall xyloglucans in Avena coleoptiles. Plant Cell Physiol 25: 1341-1351 [Google Scholar]
- Johansson P, Denman S, Brumer H, Kallas AM, Henriksson H, Bergfors T, Teeri TT, Jones TA (2003) Crystallization and preliminary analysis of a xyloglucan endotransglycosylase from Populus tremula × tremuloides. Acta Crystallogr Sect D-Biol Crystallogr 59: 535-537 [DOI] [PubMed] [Google Scholar]
- Kato Y, Ito S, Iki K, Matsuda K (1982) Xyloglucan and β-d-glucan in cell walls of rice seedlings. Plant Cell Physiol 23: 351-364 [Google Scholar]
- Moore PJ, Staehelin LA (1988) Immunogold localization of the cell-wallmatrix polysaccharides rhamnogalacturonan I and xyloglucan during cell expansion and cytokinesis in Trifolium pratense L.: implication for secretory pathways. Planta 178: 353-366 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yokoyama R, Tomita E, Nishitani K (2003) Two azuki bean XTH genes, VaXTH1 and VaXTH2, with similar tissue-specific expression profiles, are differently regulated by auxin. Plant Cell Physiol 44: 16-24 [DOI] [PubMed] [Google Scholar]
- Nishitani K (1997) The role of endoxyloglucan transferase in the organization of plant cell wall. Int Rev Cytol 173: 157-206 [DOI] [PubMed] [Google Scholar]
- Nishitani K, Masuda Y (1981) Auxin-induced changes in the cell wall structure: changes in the sugar compositions, intrinsic viscosity and molecular weight distributions of matrix polysaccharides of the epicotyls cell wall of Vigna angularis. Physiol Plant 52: 482-494 [Google Scholar]
- Nishitani K, Tominaga R (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058-21064 [PubMed] [Google Scholar]
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, Nishitani K (1993) Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J Biol Chem 268: 25364-25368 [PubMed] [Google Scholar]
- Palmer SJ, Davies WJ (1996) An analysis of relative elemental growth rate, epidermal cell size and xyloglucan endotransglycosylase activity through the growing region of ageing maize leaves. J Exp Bot 47: 339-347 [Google Scholar]
- Pritchard J, Hetehrington PRE, Fry SC, Tomos AD (1993) Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. J Exp Bot 44: 1281-1289 [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421-1435 [DOI] [PubMed] [Google Scholar]
- Rose JKC, O'Neill MA, Albersheim P, Darvill AG (2000) Functions of the plant primary cell wall. In B Ernst, G Hart, P Sinay, eds, Oligosaccharides in Chemistry and Biology, Vol II, Biology of Saccharide. Pub Wiley/VCH, Weinheim, Germany, pp 783-806
- Saab IN, Sachs MM (1996) A flooding-induced xyloglucan endotransglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiol 112: 385-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Burr B (2000) International rice genome sequencing project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3: 138-141 [DOI] [PubMed] [Google Scholar]
- Smith RC, Matthews PR, Schunmann PHD, Chandler PM (1996) The regulation of leaf elongation and xyloglucan endotransglycosylase by gibberellin in Himalay barley (Hordeum vulgare L.). J Exp Bot 47: 1395-1404 [Google Scholar]
- Tabuchi A, Mori H, Kamisaka S, Hoson T (2001) A new type of endoxyloglucan transferase devoted to xyloglucan hydrolysis in the cell wall of azuki bean epicotyls. Plant Cell Physiol 42: 154-161 [DOI] [PubMed] [Google Scholar]
- Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M (2000) Characterization of XET-related genes of rice. Plant Physiol 122: 853-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen J-P, Miller JG, Fry SC (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12: 1229-1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Verbelen J-P, Fry SC (2001) Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiol 127: 1125-1135 [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC (1994) Root growth maintenance at low water potentials. Plant Physiol 106: 607-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J (1996) The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J 9: 879-889 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2000) Functional diversity of xyloglucan-related proteins and its implications in the cell wall dynamics in plants. Plant Biol 2: 598-604 [Google Scholar]
- Yokoyama R, Nishitani K (2001a) Endoxyloglucan transferase is localized both in the cell plate and in the secretory pathway destined for the apoplast in tobacco cells. Plant Cell Physiol 42: 292-300 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2001b) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025-1033 [DOI] [PubMed] [Google Scholar]