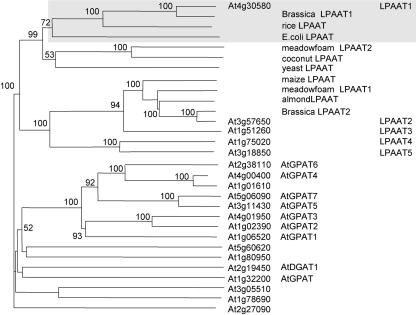
Figure 1.
A phylogenetic tree of Arabidopsis genes that encode proteins related to LPAAT constructed on the basis of their predicted amino acid sequences. It was inferred from the alignment using the neighbor-joining method with 1,000 bootstrap replicates. Only bootstrap values of over 50% are shown. Genes encoding these proteins were obtained after a BLAST search of the databases of The Arabidopsis Information Resource and National Center for Biotechnology Information with the use of the amino acid sequences of a maize cytoplasmic LPAAT (Z29518) and a B. napus plastid LPAAT1 (AF111161) as queries (for details, see “Results”). All of the preceding and following numbers of genes/proteins are from GenBank. Eleven putative LPAATs cited in a Web site (Beisson et al., 2003) and studied ATs (GPAT and DGAT1) of the Kennedy pathway are included. All reported LPAATs of other plant species (rice, AC068923; meadowfoam LPAAT2, S60477; coconut, U29657; meadowfoam LPAAT1, S60478; almond, AF213937; and B. napus LPAAT2, Z95637) and Escherichia coli (from plsC, M63491) and yeast (Saccharomyces cerevisiae) LPAAT (from slc1, L13282) are incorporated. The Arabidopsis genes are shown by their locus numbers, and the first five genes are also shown by their simplified protein names, as are those from other plants and microbes. Arabidopsis LPAAT1, B. napus plastid LPAAT1, a rice LPAAT (presumably in the plastids), and E. coli LPAAT are shaded.