Abstract
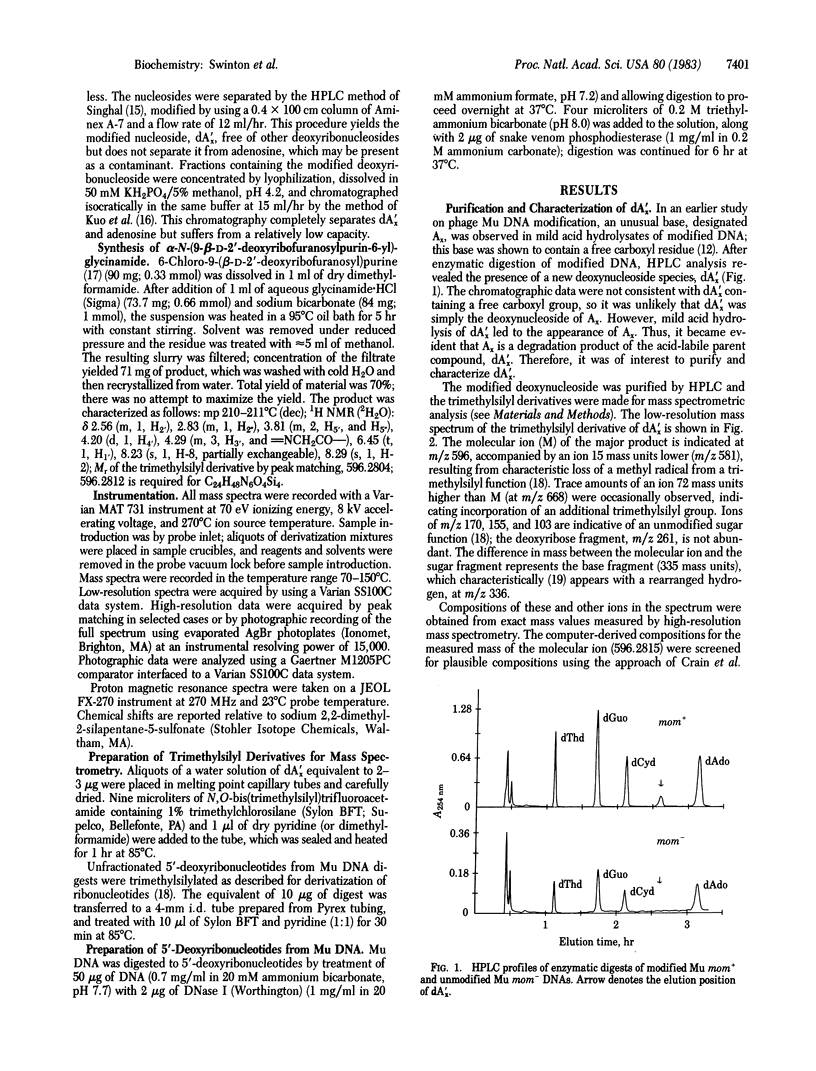
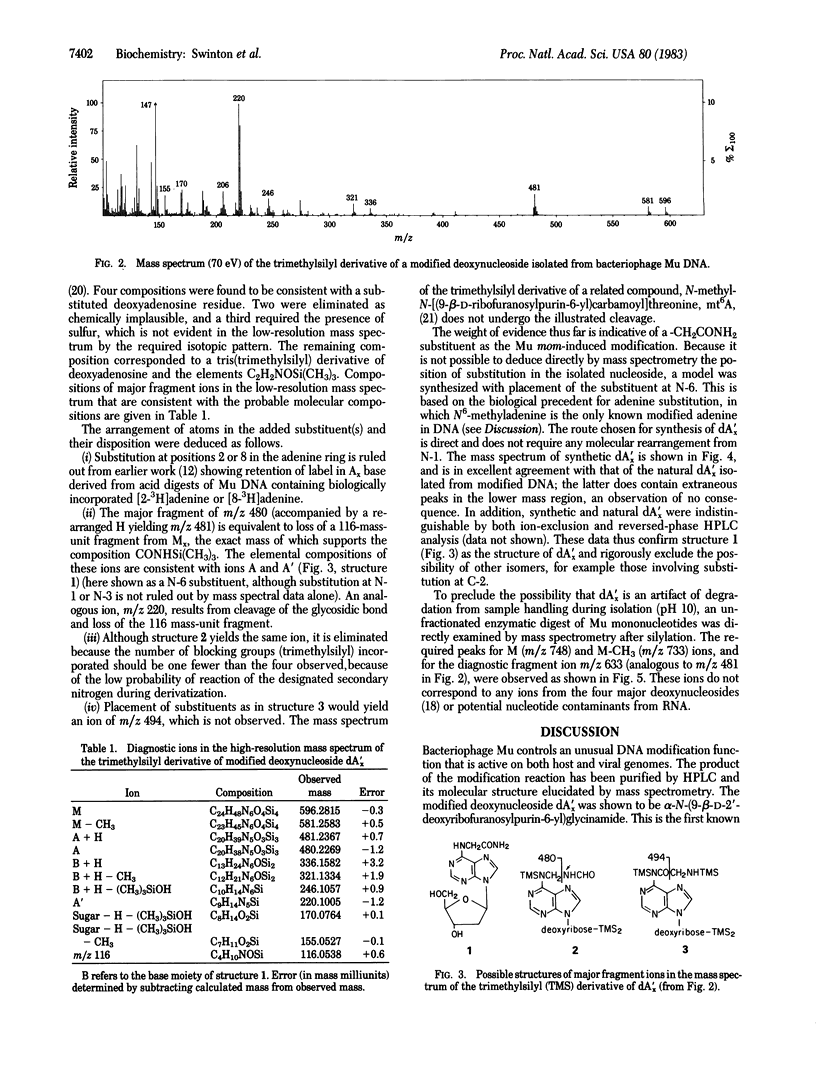
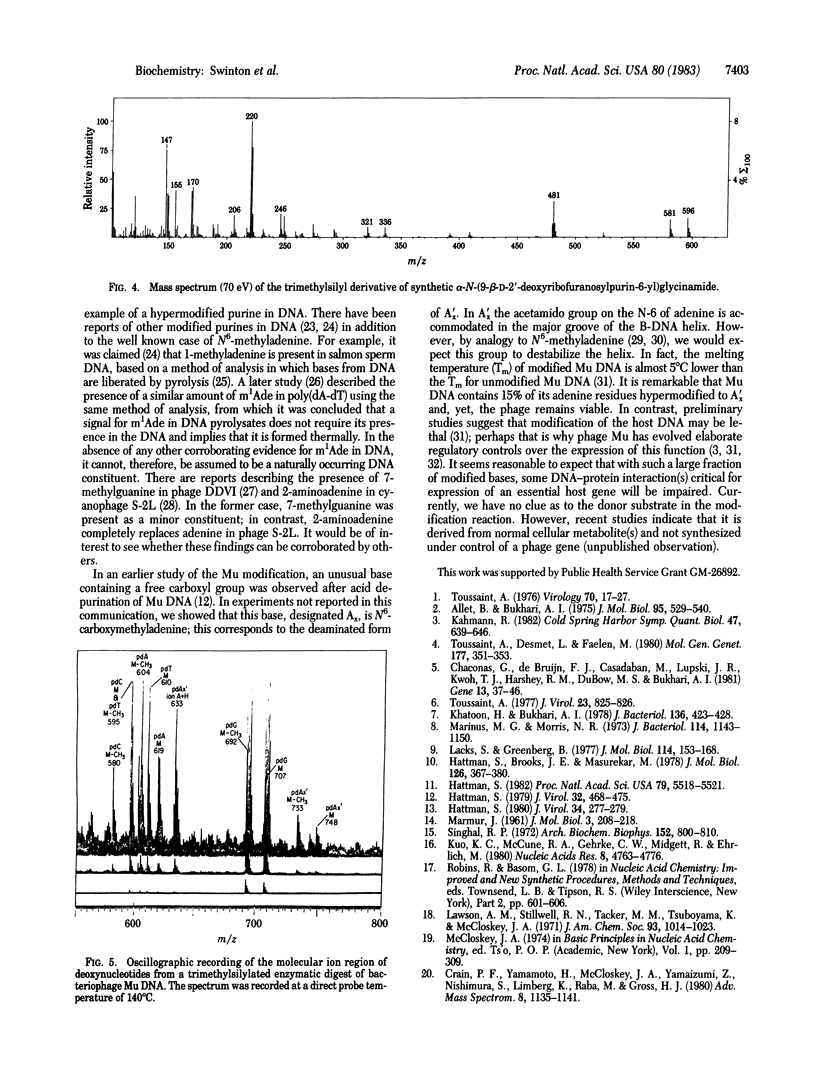
Bacteriophage Mu encodes a protein that modifies approximately equal to 15% of DNA adenine residues to a new and unusual form. Modified DNA was enzymatically digested to deoxynucleosides, and the products were fractionated by HPLC. A modified adenine nucleoside, designated dA'x, was purified and its molecular structure was established by mass spectrometry. We show that dA'x is alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)-glycinamide. The dA'x obtained from DNA was indistinguishable from the synthetic product with respect to its chromatographic behavior (HPLC and gas chromatography) and mass spectrum. Acid hydrolysis degrades dA'x to produce N6-carboxymethyladenine; this compound corresponds to the base Ax observed in earlier studies.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Chaconas G., de Bruijn F. J., Casadaban M. J., Lupski J. R., Kwoh T. J., Harshey R. M., DuBow M. S., Bukhari A. I. In vitro and in vivo manipulations of bacteriophage Mu DNA: cloning of Mu ends and construction of mini-Mu's carrying selectable markers. Gene. 1981 Jan-Feb;13(1):37–46. doi: 10.1016/0378-1119(81)90041-x. [DOI] [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J Biol Chem. 1978 Feb 10;253(3):927–934. [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations: studies at the monomer level. Biochemistry. 1974 Sep 24;13(20):4143–4158. doi: 10.1021/bi00717a013. [DOI] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S. DNA methyltransferase-dependent transcription of the phage Mu mom gene. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5518–5521. doi: 10.1073/pnas.79.18.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Goradia M., Monaghan C., Bukhari A. I. Regulation of the DNA-modification function of bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):647–653. doi: 10.1101/sqb.1983.047.01.076. [DOI] [PubMed] [Google Scholar]
- Hattman S. Specificity of the bacteriophage Mu mom+ -controlled DNA modification. J Virol. 1980 Apr;34(1):277–279. doi: 10.1128/jvi.34.1.277-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Unusual modification of bacteriophage Mu DNA. J Virol. 1979 Nov;32(2):468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):639–646. doi: 10.1101/sqb.1983.047.01.075. [DOI] [PubMed] [Google Scholar]
- Khatoon H., Bukhari A. I. Bacteriophage Mu-induced modification of DNA is dependent upon a host function. J Bacteriol. 1978 Oct;136(1):423–428. doi: 10.1128/jb.136.1.423-428.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Harada F., Von Minden D. L., McCloskey J. A., Nishimura S. N-((9- -D-ribofuranosylpurin-6-yl)-N-methylcarbamoyl) threonine, a modified nucleoside isolated from Escherichia coli threonine transfer ribonucleic acid. Biochemistry. 1972 Oct 10;11(21):3910–3915. doi: 10.1021/bi00771a012. [DOI] [PubMed] [Google Scholar]
- Kirnos M. D., Khudyakov I. Y., Alexandrushkina N. I., Vanyushin B. F. 2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature. 1977 Nov 24;270(5635):369–370. doi: 10.1038/270369a0. [DOI] [PubMed] [Google Scholar]
- Kuo K. C., McCune R. A., Gehrke C. W., Midgett R., Ehrlich M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 1980 Oct 24;8(20):4763–4776. doi: 10.1093/nar/8.20.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lawson A. M., Stillwell R. N., Tacker M. M., Tsuboyama K., McCloskey J. A. Mass spectrometry of nucleic acid components. Trimethylsilyl derivatives of nucleotides. J Am Chem Soc. 1971 Feb 24;93(4):1014–1023. doi: 10.1021/ja00733a039. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Debov S. S. Methylated guanine derivative as a minor base in the DNA of phage DDVI Shigella disenteriae. Biochim Biophys Acta. 1976 Jun 18;435(2):206–210. doi: 10.1016/0005-2787(76)90251-3. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vrieling H., Van de Putte P. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-coded transactivator. Nature. 1983 Jan 27;301(5898):344–347. doi: 10.1038/301344a0. [DOI] [PubMed] [Google Scholar]
- Schoen A. E., Cooks R. G., Wiebers J. L. Modified bases characterized in intact DNA by mass-analyzed ion kinetic energy spectrometry. Science. 1979 Mar 23;203(4386):1249–1251. doi: 10.1126/science.570725. [DOI] [PubMed] [Google Scholar]
- Singhal R. P. Ion-exlusion chromatography: analysis and isolation of nucleic acid components, and influence of separation parameters. Arch Biochem Biophys. 1972 Oct;152(2):800–810. doi: 10.1016/0003-9861(72)90276-7. [DOI] [PubMed] [Google Scholar]
- Toussaint A. DNA modification of bacteriophage Mu-1 requires both host and bacteriophage functions. J Virol. 1977 Sep;23(3):825–826. doi: 10.1128/jvi.23.3.825-826.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A., Desmet L., Faelen M. Mapping of the modification function of temperate phage Mu-1. Mol Gen Genet. 1980 Jan;177(2):351–353. doi: 10.1007/BF00267450. [DOI] [PubMed] [Google Scholar]
- Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976 Mar;70(1):17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- Wiebers J. L. Detection and identification of minor nucleotides in intact deoxyribonucleic acids by mass spectrometry. Nucleic Acids Res. 1976 Nov;3(11):2959–2970. doi: 10.1093/nar/3.11.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]


