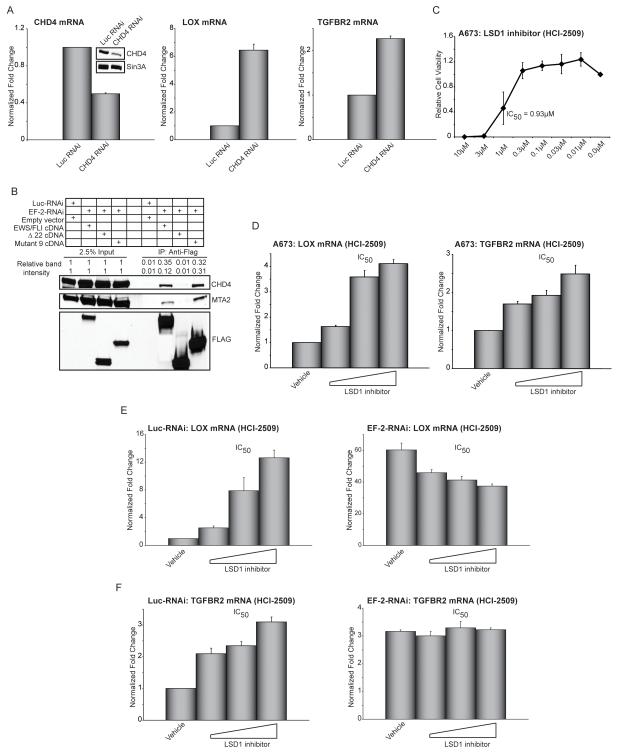
Figure 6. EWS/FLI interacts with members of the NuRD co-repressor complex.
(A) qRT-PCR analysis of CHD4, LOX, and TGFBR2 in A673 cells following knock-down of the NuRD component CHD4. Western blot analysis to demonstrate efficiency of CHD4 knockdown, Sin3A was used as the loading control. Error bars indicate standard deviations. Normalized fold enrichment was calculated by determining the fold-change of each condition relative to the control Luc-RNAi condition, with the data in each condition normalized to an internal housekeeping control gene GAPDH.
(B) Co-immunoprecipitation of 3X-FLAG EWS/FLI, or the indicated mutants, and NuRD complex components CHD4 and MTA2. Relative band intensities were quantified using ImageQuant.
(C) Relative cell viability of A673 cells treated with increasing concentrations of the LSD1 inhibitor (HCI-2509). The IC50 is indicated. Error bars indicate standard deviations.
(D) qRT-PCR analysis of LOX and TGFBR2 following 72 hours of treatment with increasing concentrations of the LSD1 inhibitor (HCI-2509). The dose corresponding to the IC50 is indicated. Error bars indicate standard deviations.
(E, F) qRT-PCR analysis of LOX and TGFBR2 in A673 cells expressing Luc-RNAi (negative control) or EWS/FLI RNAi (EF-2-RNAi) following 72 hours of treatment with increasing concentrations of the LSD1 inhibitor HCI-2509. The dose corresponding to the IC50 is indicated. Error bars indicate standard deviations.