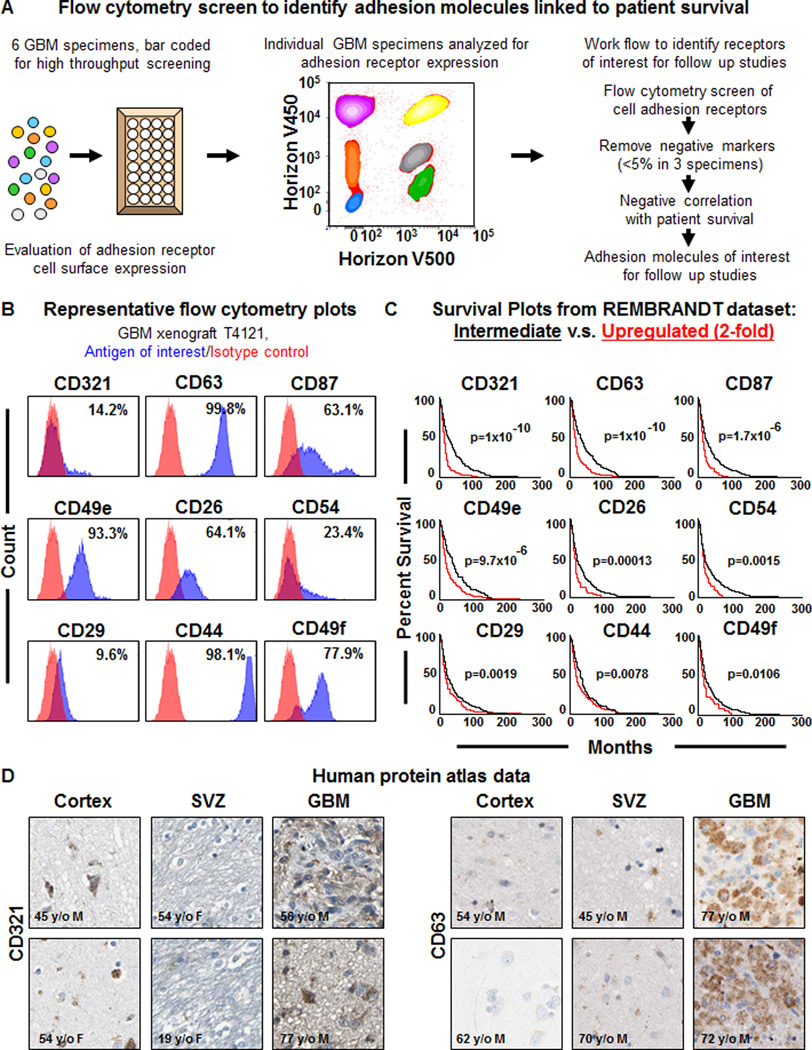
Figure 1. High throughput screening on GBM specimens using a multiplex flow cytometry identifies cell adhesion receptors negatively correlated to GBM patient survival.
Schematics depicting the work flow of the high throughput flow cytometry screening method (A) based on bar coding multiple GBM specimens and screening cell surface marker expression to identify cell adhesion receptors linked to GBM patient survival. Representative flow cytometry plots (B) for adhesion receptors from GBM xenograft T4121, antigen of interest shown in blue, isotype control shown in red, and percentage of positive cells indicated on each individual plot. Representative Kaplan-Meier survival curves (C) for candidate adhesion molecular generated from the NCI REpository for Molecular BRAin Neoplasia DaTa (REMBRANDT) bioinformatics database (https://caintegrator.nci.nih.gov/rembrandt/) for the 2-fold upregulated (red) and intermediate (black) groups. Log-rank p-value for survival curves between upregulated and intermediate groups provided for each adhesion molecule. Representative immunohistochemical data (D) generated for CD321 and CD63 from the human protein atlas (http://www.proteinatlas.org) for the cortex, subventricular zone (SVZ), and glioblastoma (GBM) tumor. Age and sex of each individual indicated directly on the micrographs. See also Table S1, Figure S1.