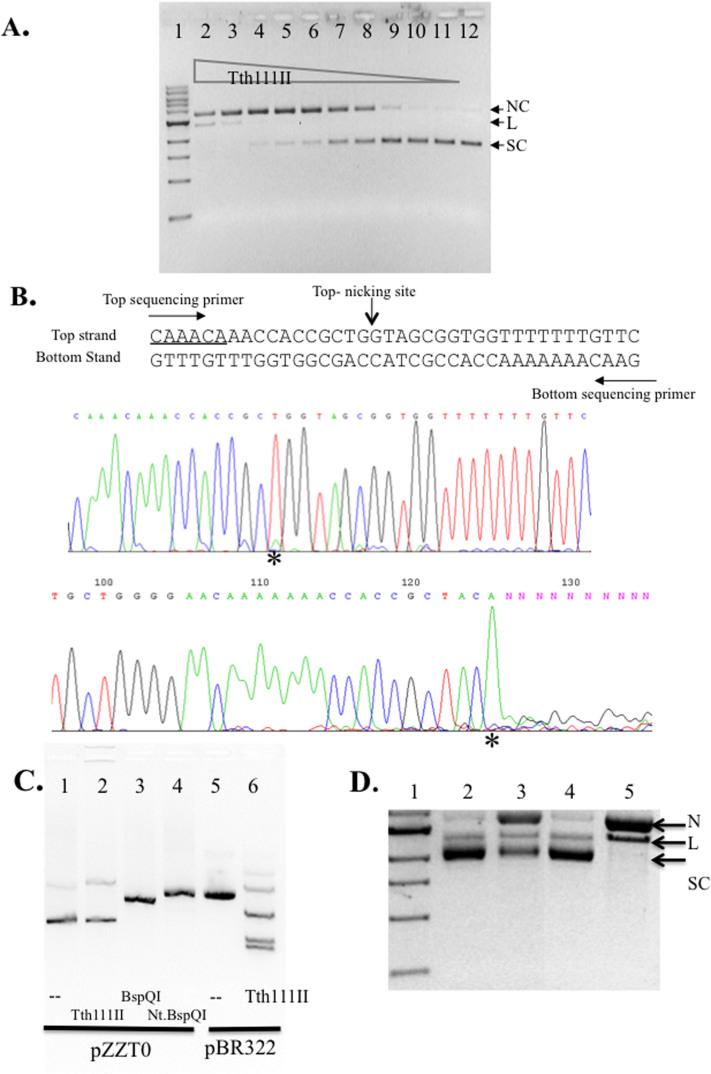
Figure 4. Tth111II activity assay on a single-site substrate and run-off sequencing of the nicked products.
(A). Tth111II digestion of pZZT1 with a single Tth111II site plus 320 μM SAM. Lane 2–11; 220 ng pZZT1 was digested with Tth111II in a 2-fold enzyme serial dilution (lane 2 enzyme = 91 pmole); Lane 12, uncut pZZT1. (B). Run-off sequencing of the nicked pZZT1. The template sequence and the sequencing primers and nicking sites are shown. Top sequencing panel, sequence read from the top sequencing primer. Bottom sequencing panel, sequence read from bottom sequencing primer. “*” indicates extra “A” peak in the run-off reactions. (C). Digestion of pZZT0 (no Tth111II site) by Tth111II endonuclease. (D). Stepwise methylase and endonuclease activity assays for RM.Tth111II in the absence of or presence of Mg2+. Lane 1, DNA marker; lanes 2, partially modified pZZT1 after methylation in the digestion buffer in the absence of Mg2+; lane 3, endonuclease digestion of the partially modified pZZT1 in the presence of Mg2+ and SAM (note, the remaining supercoiled DNA is resistant to digestion/nicking due to the methylation of Tth111II site. Thus the remaining supercoiled DNA is an indication of methylase activity of RM.Tth111II); lane 4, Mock-modified (unmethylated) pZZT1; lane 5, mock-modified pZZT1 DNA digested by Tth111II in the presence of Mg2+. N, nicked circular; L, linear DNA; S, supercoiled DNA.