Abstract
Strategies to detect human DNA methyltransferases are needed, given that aberrant methylation by these enzymes is associated with cancer initiation and progression. Here we describe a non-radioactive, antibody-free, electrochemical assay in which methyltransferase activity on DNA-modified electrodes confers protection from restriction for signal-on detection. We implement this assay with a multiplexed chip platform and show robust detection of both bacterial (SssI) and human (Dnmt1) methyltransferase activity. Essential to work with human methyltransferases, our unique assay design allows activity measurements on both unmethylated and hemimethylated DNA substrates. We validate this assay by comparison with a conventional radioactive method. The advantages of electrochemistry over radioactivity and fluorescence make this assay an accessible and promising new approach for the sensitive, label-free detection of human methyltransferase activity.
INTRODUCTION
In mammals, DNA methylation is the most prominent form of epigenetic gene regulation and is a critical long-term gene silencing mechanism.1,2 This covalent addition of a methyl group to the carbon-5 position of cytosine at predominantly 5′-CG-3′ sites is catalyzed by DNA methyltransferases, which use the cofactor S-adenosyl-L-methionine (SAM) as a methyl donor. DNA methylation is central to many normal cellular processes including development, X chromosome inactivation, gene regulation, and transposon silencing, among others.1,2 However, aberrant DNA methylation has been associated with multiple disease states including developmental abnormalities such as ICF (immunodeficiency, centromere instability, and facial abnormalities) syndrome and Rett syndrome,3,4 autoimmune diseases such as lupus,5 and many types of cancer.6–8
The link between abnormal DNA methylation and cancer has recently become an area of intense, widespread research, and both excessive methylation (hypermethylation) and deficient methylation (hypomethylation) have been identified in diverse tumor types.7,9 While hypermethylation can contribute to oncogenesis by the silencing of tumor suppressor genes,8 hypomethylation may activate oncogenes or latent retrotransposons, or cause chromosome instability.7 In many cases, these harmful methylation states have been linked to the abnormal expression and activity of DNA methyltransferases.8,10–13
Mammalian DNA methyltransferases include Dnmt1, Dnmt3a, and Dnmt3b and while all three catalyze the same reaction, they play different roles in establishing methylation patterns in the genome. Dnmt1 transmits methylation patterns across cell divisions by completing methylation on newly replicated strands at 5′-CG-3′ sites that carry methylation on the template strand alone.1 Thus Dnmt1 is characterized as a maintenance methyltransferase and displays a significant preference for hemimethylated DNA substrates.14 Dnmt3a and Dnmt3b, in contrast, are characterized as de novo methyltransferases because of their activity at unmethylated 5′-CG-3′ sites, primarily during embryogenesis when new methylation patterns must be set.1,15 These inherently different activities contribute to the complex roles of methyltransferases that are now being elucidated in a growing number of cancers.
Understanding how methyltransferase activity contributes to cancer initiation and progression is a very attractive goal; abnormalilties in methyltransferase activity usually occur far before other signs of malignancy and could thus be used for early cancer detection.7,8 Additionally, identification of cancers with a certain methylation phenotype (hypermethylation or hypomethylation) can help specify an effective course of treatment.7,16 Clearly, the expansion of this new field, including the study of methyltransferase activity in cancer cells, the characterization of anti-methylation drugs, and the screening of patients for early cancer diagnosis, requires effective and accessible assays to measure methyltransferase activity.
While radioactive labeling with [methyl-3H]-SAM is the current standard for assaying methyltransferase activity,14,17 the desire to avoid radioactive reagents has motivated the development of diverse alternatives including PCR-based bisulfite conversion,18 HPLC,19 and fluorescence and colorimetric assays.20,21 Additionally, assays have been developed that utilize digestion of DNA by methylation-sensitive restriction enzymes. In these assays, methylation of a specific DNA sequence confers protection from digestion by the corresponding restriction enzyme, and results are typically visualized by fluorescence.22–24 Alhough non-radioactive, these methods still carry significant drawbacks including time-consuming sample preparation and data analysis, bulky detection equipment, expensive antibodies and fluorescently labeled substrates, and inflexible detection schemes that are not compatible with human methyltransferases.
Electrochemical strategies overcome many of these drawbacks, providing non-radioactive, low-cost, portable sensors that have high potential for use in clinical settings.25,26 Despite these advantages, relatively little work has been aimed at the electrochemical detection of methyltransferase activity. Reported electrochemical strategies include the direct oxidation of individual DNA bases to detect 5-methylcytosine27 and several methods that use methylation-sensitive restriction enzymes. These include restriction-based signal modulation with DNA-functionalized gold nanoparticles,28 restriction-facilitated binding of redox-active moieties such as carbon nantubes,29 probe-modified DNA,30 and redox-active enzymes,31 and DNA monolayers with methylation-sensitive restriction sites that bear either electrochemical32–35 or photoelectrochemical36 reporters. Though diverse, these strategies are limited in that they are either demonstrated with synthetic 5-methylcytosine alone and not enzymatic methylation or they are only applicable to the detection of bacterial methyltransferase activity. As human methyltransferase activity is sensitive to the methylation state of the DNA substrate (unmethylated or hemimethylated), assays for the study of human methyltransferases must allow for the use of both substrates.
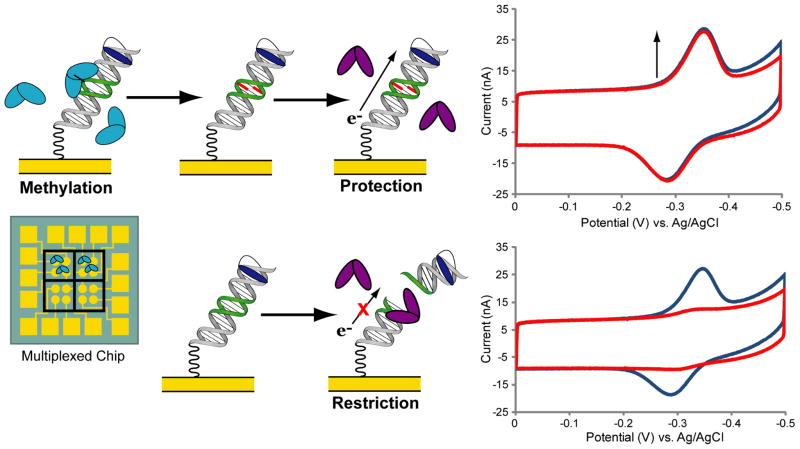
Here we describe a new electrochemical assay in which either an unmethylated or hemimethylated DNA substrate may be used for the sensitive detection of both bacterial and human methyltransferase activity. In this assay, multiplexed, DNA-modified electrodes bearing covalent redox probes26,37 are combined with a methylation-sensitive restriction enzyme to convert methylation into an electrical signal (Figure 1). Such DNA-modified electrodes have been used previously to detect protein binding38 and restriction activity,37–39 and the electrochemistry of the covalent methylene blue redox probe employed here has been thoroughly characterized.40 Importantly, our multiplexed chip platform (Figure 1, inset) allows for the direct comparison of up to four types of DNA substrates and four methyltransferase conditions side by side on the same surface.37 For this assay, electrodes are modified with DNA that contains the human methylation site (5′-CG-3′) within the recognition site of a methylation-sensitive restriction enzyme. Upon treatment of the electrodes with active methyltransferases, these sites become methylated, thereby protecting the DNA from restriction during subsequent restriction enzyme treatment. With the DNA intact, the redox signal from the probe is retained (signal-on). In the absence of active methyltransferases, the DNA remains unmethylated and is readily cut, causing the near complete disappearance of the redox signal (signal-off).
Figure 1.
Assay for the electrochemical detection of methyltransferase activity. DNA-modified electrodes with overlapping recognition sites of a methyltransferase and restriction enzyme (green section of DNA) are prepared on multiplexed chips (inset). In the presence of active methyltransferases (top left), the DNA-modified electrodes become methylated (red DNA bases) and protected from cutting during subsequent restriction enzyme treatment. Thus by cyclic voltammetry (CV) scans (top right), the covalent methylene blue redox probe exhibits a signal-on result both before (blue traces) and after (red traces) restriction enzyme treatment. In the absence of active methyltransferases (bottom left), the DNA remains unmethylated and is readily cut by the restriction enzyme. This signal-off result is reported (bottom right) by signal attenuation upon restriction enzyme treatment. Multiplexed chips consist of 16 individually addressed electrodes divided into quadrants of 4 electrodes. Each quadrant may be modified with a different DNA substrate and treated with a unique methyltransferase sample, allowing for the analysis of up to four DNA substrates and four methyltransferase conditions side by side on the same device.
Importantly, this work addresses the special requirements and challenges associated with human methyltransferase detection. First, DNA substrate versatility is critical as the primary human methyltransferase, Dnmt1, has a strong preference for hemimethylated 5′-mCG-3′ sites. To meet this requirement, we demonstrate this assay with both the BstUI and BssHII restriction endonucleases (recognition sites of 5′-CGCG - 3′ and 5′-GCGCGC-3′, respectively). While BstUI does not support the use of a hemimethylated substrate because hemimethylation of its recognition site alone blocks restriction, BssHII allows for the use of both unmethylated (5′-GCGCGC-3′) and hemimethylated (5′-GmCGCGC-3′) DNA because both substrates are readily cut if not further methylated. Second, work with human methyltransferases involves the exposure of electrode surfaces to greater amounts of protein material due to the larger size and lower activity of these proteins, as compared to bacterial methyltransferases. To overcome the obstructive effects of high protein content on electrochemical signals, we introduce a simple and effective protease treatment step. With these important adaptations we are able, uniquely, to detect human methyltransferase activity by an electrochemical method. We demonstrate this assay for the sensitive detection of bacterial SssI and human Dnmt1 methyltransferase activity with a multiplexed, low cost format that may easily be applied to high throughput studies or utilized in research and clinical laboratories.
MATERIALS AND METHODS
Materials
All standard and modified phosphoramidites were purchased from Glen Research. Modified methylene blue dye for coupling was synthesized as described previously.40 S-adenosyl-L-methionine (SAM) and lambda DNA were purchased from New England Biolabs. Tritiated SAM (32H-SAM) was purchased from Perkin Elmer. All other chemicals for the preparation of protein buffers and DNA-modified electrodes, and for use in 32H-SAM experiments were purchased from Sigma-Aldrich and used as received. Multiplexed chips were fabricated at Caltech as described previously.37
Protein Preparation
All proteins were purchased from commercial sources. SssI methyltransferase, BSA, and the restriction endonucleases BstUI, BssHII, and RsaI were purchased from New England Biolabs and used as received unless otherwise indicated. Protease from Streptomyces griseus was purchased as a dry powder from Sigma-Aldrich and stored as a 250 μM solution in 40% glycerol in phosphate buffer without NaCl (5 mM phosphate, pH 7) at −20°C. Human Dnmt1 was purchased from BPS Bioscience. Buffer exchange by size exclusion spin column (10 kDa cutoff, Amicon) was performed on Dnmt1 and BssHII prior to electrochemistry experiments to remove dithiothreitol (DTT), which disrupts DNA-modified electrodes upon heating. The exchange was performed according to manufacturer instructions at 4°C. Dnmt1 was exchanged into Dnmt1 activity buffer (50 mM Tris-HCl, 1 mM EDTA, 5% Glycerol, pH 7.8), while BssHII was exchanged into a methylation/restriction (M/R) buffer (10 mM tris-HCl, 50 mM NaCl, 10 mM MgCl2, pH 7.9)
DNA Sequences
For the detection of methyltransferase activity with BstUI, electrodes were modified with the sequence 5′-HS- (CH2)6 -GACTGAGTACTCGCGACTGA -3′ with an unmethylated methylene blue-modified complement. The BstUI restriction site (5′-CGCG - 3′) is underlined. As a control, this sequence also contains the RsaI restriction site (5′-GTAC-3′) which is italicized. For experiments with synthetically methylated DNA, the BstUI restriction site was fully methylated on both strands (5′-mCGmCG -3′).
For the detection of methyltransferase activity with BssHII, electrodes were modified with the sequence 5′-HS- (CH2)6 - GACTGAGTACTGCGCGCACTGA -3′ with an unmethylated methylene blue-modified complement. The unmethylated BssHII restriction site (5′-GCGCGC -3′) is underlined. DNA was also prepared with a hemimethylated BssHII restriction site (5′-GmCGCGC -3′).
DNA Synthesis
All DNA was synthesized with an Applied Biosystems 3400 DNA synthesizer. Thiolated strands were prepared with a C6-S-S phosphoramidite at the 5′ terminus. Strands containing methylated cytosine were synthesized with a 5-methyl dC-CE phosphoramidite. DNA for methylene blue coupling was prepared with an amino-C6-dT phosphoramidite at the 5′ terminus. All DNA was purified by reverse-phase HPLC with a polymeric PLRP-S column (Agilent) and characterized by mass spectrometry. For methylene blue-modified DNA, coupling was carried out in solution as described previously.40 Briefly, HPLC-purified DNA was suspended in 0.1 M NaHCO3 and combined with an equimolar amount of modified methylene blue dye in DMSO. The mixture was allowed to shake overnight at room temperature. The coupled DNA was then purified by Nap-5 size exclusion column (GE Healthcare) before further purification by HPLC. For thiolated DNA, the disulfide was reduced to the free thiol with 100 mM DTT in 100 mM tris-HCl buffer, pH 8.3 at room temperature for 45 minutes, and then purified by Nap-5 column and HPLC. To prepare duplexes, all DNA stocks were desalted, resuspended in phosphate buffer (5 mM phosphate, 50 mM NaCl, pH 7), and quantified by UV/Vis absorption at 260 nm. Equimolar amounts (50 μM) of complementary strands were combined and thermally annealed.
Multiplexed Chip Preparation and Assembly
Prior to application of DNA solutions, chips were cleaned with acetone and isopropanol, dried, and further cleaned by UV ozone. The chips were then assembled with a rubber gasket and clamp, and a solution of 25 μM DNA in phosphate buffer was immediately applied (20 μL DNA per quadrant). DNA film assembly was allowed to proceed overnight at room temperature in a humid environment.
Electrochemistry
All electrochemistry was carried out with a standard potentiostat and multiplexer console (CH Instruments). A three-electrode system was employed including a Pt wire auxiliary electrode and Ag/AgCl reference electrode (Cypress Systems). DNA-modified chips were first backfilled with 1 mM mercaptohexanol in phosphate buffer with 5% glycerol for 45 minutes at room temperature. For all electrochemistry, cyclic voltammetry (CV) scans were performed at a 100 mV/s scan rate over a potential window of 0 mV to −500 mV. Scans of the 16 electrodes on a chip were performed sequentially. Following each treatment step, chips were rinsed and scanned with 200 μL of the specified buffer in a common well. Signal size was measured as the CV cathodic peak area. The reported variation in the data represents the standard deviation across all electrodes measured for a given condition.
For the detection of SssI activity, scans were performed in M/R buffer. Individual quadrants of chips were treated with SssI in M/R buffer with 160 μM SAM. A reaction volume of 10 μL was used for each quadrant. The SssI solution was allowed to incubate on the chips at room temperature for 2 hours. Chips were then rinsed and treated with a solution of 10 μg/mL lambda DNA in M/R buffer for 45 minutes at room temperature. Following this, chips were rinsed with 500 mM NaCl in M/R buffer to ensure complete SssI dissociation. Next, chips were treated with 1,000 units/mL of BstUI in M/R buffer at room temperature for 1 hour. As a control, chips were then treated with 1,000 units/mL RsaI in M/R buffer at room temperature for 30 minutes.
A similar procedure was followed for the detection of Dnmt1. As Dnmt1 activity buffer lacks ionic and charged components (such as NaCl, MgCl2, and spermidine which inhibit Dnmt1 activity),41 it is not optimal for DNA electrochemistry. Thus, after Dnmt1 methylation was allowed to proceed in Dnmt1 activity buffer, an optimized scanning buffer (5 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 μM EDTA, 10% glycerol, pH 7) was used for all electrochemical scans. Dnmt1 with 100 μg/mL BSA and 160 μM SAM were applied to individual chip quadrants and chips were incubated at 37°C for 2 hours in a humidified container. Chips were then treated with 1 μM protease in phosphate buffer for 1 hour at 37°C. After thorough rinsing, chips were treated with 1,500 units/mL of BssHII in M/R buffer at 37°C for 1 hour. For measurements of SssI activity on the BssHII 22-mer, scans were also performed in optimized scanning buffer and protease treatment was used. Including the methyltransferase and restriction enzyme incubations, the total assay time for SssI or Dnmt1 is about 5 hours.
32H-SAM Methyltransferase Activity Assay
Methyltransferase activity was also determined by a conventional 32H-SAM activity assay, based partially on previously published procedures.14,17 Briefly, 20 μL reactions were prepared with 20 μM DNA, 0.5 μCi 32H-SAM (~3 μM), and the methyltransferase sample in appropriate activity buffer. For reactions with Dnmt1, 100 μg/mL BSA was included. For the DNA substrate, the same BssHII unmethylated and hemimethylated 22-mer sequence used for electrochemistry was employed, but without any probe or thiol modifications. For each experiment, positive (15 nM SssI) and negative (no protein) controls were included. The reactions were mixed thoroughly and incubated at 37°C for 2 hours. The reactions were then stopped with 30 μL of a 10 % TCA solution, spotted onto DE81 filter paper (Whatman), and allowed to air dry for 15 minutes. The filter papers were then soaked separately in 10 mL of 50 mM Na2HPO4 for 15 minutes, and then rinsed with 50 mM Na2HPO4 and 95% cold ethanol. The filter papers were then dried before measurement by liquid scintillation counting.
RESULTS AND DISCUSSION
Electrochemical Detection of SAM-dependent SssI Methyltransferase Activity
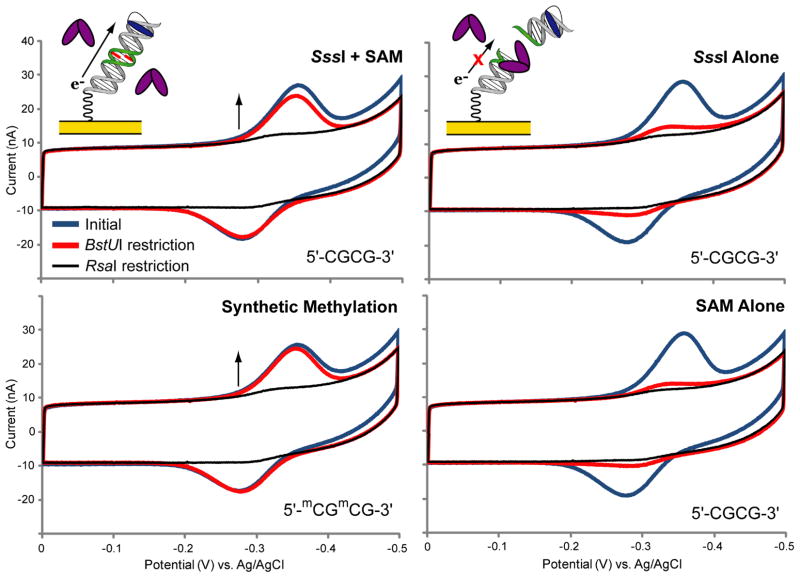
We first established this assay for the detection of SssI methyltransferase, the bacterial analog to human methyltransferases, which also binds the site 5′-CG-3′ and methylates the C-5 position of the target cytosine.42 To confirm that electrochemical signal protection is due exclusively to DNA methylation, the dependence of SssI-mediated protection on the essential SAM cofactor was evaluated. A 20-mer DNA duplex with a covalent methylene blue redox probe at one end was designed with a centrally located BstUI restriction site (5′-CGCG - 3′) and an adjacent RsaI restriction site (5′-GTAC - 3′). Importantly, while SssI can methylate the BstUI site, it does not recognize the RsaI site. Multiplexed chips were modified in three quadrants with the unmethylated BstUI 20-mer and treated side by side with 15 nM SssI with SAM, 15 nM SssI alone, or SAM alone. As a positive control, the fourth quadrant was modified with the synthetically methylated BstUI 20-mer and left untreated (Figure 2).
Figure 2.
SssI protection from BstUI restriction is dependent on the SAM cofactor. Chips were modified in three quadrants with the unmethylated BstUI 20-mer (top row and bottom right) and in one quadrant with the synthetically methylated BstUI 20-mer (bottom left). The BstUI 20-mer consists of the sequence 5′-HS- (CH2)6 - GACTGAGTACTCGCGACTGA -3′ with an unmethylated methylene blue-modified complement. The BstUI restriction site (5′-CGCG - 3′) is underlined and the RsaI restriction site (5′-GTAC-3′) is italicized. The synthetically methylated BstUI 20-mer contains a restriction site that is fully methylated on both strands (5′-mCGmCG-3′). DNA protection was evaluated side by side on the same chip: the unmethylated DNA quadrants were treated with 20 nM SssI and 160 μM SAM (top left), 20 nM SssI alone (top right), or 160 μM SAM alone (bottom right), while the synthetically methylated quadrant was left untreated in buffer alone. After an initial CV scan (blue traces), the chip was treated in all quadrants with BstUI (1,000 units/mL) (red traces). Finally, the chip was treated in all quadrants with RsaI (1,000 units/mL) (black traces). All CV scans were performed in M/R buffer (10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, pH 7.9) with an Ag/AgCl reference electrode at a 100 mV/s scan rate.
Initially, after SssI treatment of the electrodes, large electrochemical signals are observed from the methylene blue-modified DNA film. Following BstUI treatment, electrodes treated with SssI and SAM show near complete signal protection (98% ± 2%), while those treated with SssI alone or SAM alone show the same, minimal signal protection (12% ± 1% and 13% ± 1%, respectively). Thus, the signal-on response of this assay depends on the presence of the methyl donor, SAM. Electrodes modified with the synthetically methylated BstUI 20-mer show complete signal protection (no measurable signal decrease).
To further rule out other possible modes of DNA protection, such as SssI-DNA binding or nonspecific SssI aggregation that physically blocks the BstUI restriction site, the electrodes were then treated with RsaI, a restriction enzyme that is not inhibited by 5′-CG-3′ methylation. With this treatment, the remaining redox signals for all DNA types are reduced to the same, near complete level of attenuation (4% ± 1% signal remaining for all). As the RsaI recognition site is located well within the binding footprint of SssI,43 DNA binding or aggregation by SssI that prevents access of BstUI to the DNA, would also prevent the access of RsaI. These results indicate that the observed DNA protection is due specifically to DNA methylation catalyzed by SssI.
Concentration Dependence of SssI Methyltransferase Activity
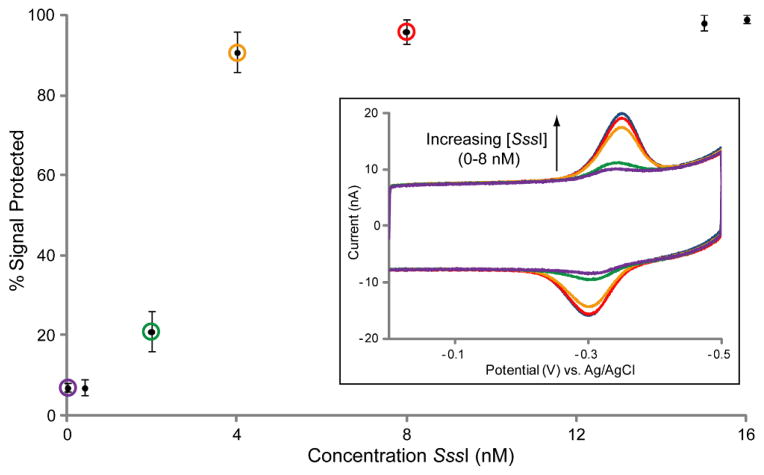
The concentration range over which SssI methyltransferase activity is detected with this assay was then evaluated. Multiplexed chips modified with the unmethylated BstUI 20-mer were treated with a range of SssI concentrations (0–16 nM) including up to four different concentrations on the same chip (Figure 3, inset). The percent signal protected from BstUI restriction at each SssI concentration, compiled from 4 chips with 4–8 electrodes measured at each concentration, was used to make an activity curve (Figure 3). Near complete signal protection is observed with SssI concentrations of 8 nM and higher (96 % ± 3% at 8 nM; 99% ± 1% at 16 nM). Between the narrow SssI concentration range of 4 nM and 2 nM, there is a sharp loss of signal protection (91% ± 5% at 4 nM; 21% ± 5% at 2 nM). Below 2 nM SssI, signal protection is not distinguished from the baseline signal that remains when electrodes are left untreated (7% ± 1%). Thus the limit of detection for SssI is 2 nM.
Figure 3.
Concentration dependence of SssI methyltransferase activity. Chips were modified in all quadrants with the unmethylated BstUI 20-mer. DNA protection by various concentrations of SssI was evaluated side by side on the same chip, and CV scans from representative electrodes are shown overlaid (inset). Overlaid CV traces include the initial signal before BstUI treatment (blue), 8 nM SssI (red), 4 nM SssI (orange), 2 nM SssI (green), and untreated (purple). Quantification of the observed signal protection is displayed as an SssI activity curve where the circled points correspond to the concentrations represented by the overlaid CV traces. Error bars represent the standard deviation across 4–8 electrodes tested for each SssI concentration on 4 chips. All CV scans were performed in M/R buffer (10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, pH 7.9) with an Ag/AgCl reference electrode at a 100 mV/s scan rate.
A Hemimethylated DNA Substrate and Protease Treatment for Human Dnmt1 Detection
To accommodate the most prominent human methyltransferase, Dnmt1, which shows strong preferential activity at hemimethylated 5′-mCG-3′ sites, DNA substrates with a hemimethylated BssHII restriction site (5′-GmCGCGC-3′) were utilized. Importantly, BssHII requires full methylation of either 5′-CG-3′ site within its recognition sequence to prevent DNA restriction. Given that Dnmt1 is larger than SssI (molecular weights of 185 kD and 42 kD, respectively) as well as less active, studies with Dnmt1 require the exposure of electrodes to solutions with greater amounts of total protein material. Upon addition of 100 nM Dnmt1 to DNA-modified electrodes, substantial broadening of the redox peak is observed along with some signal loss (Figure 4). These effects severely interfere with activity comparisons across a wide Dnmt1 concentration range. Unlike work with SssI, where binding can be reversed by competitor DNA and buffer washes, these treatments are not effective for Dnmt1.
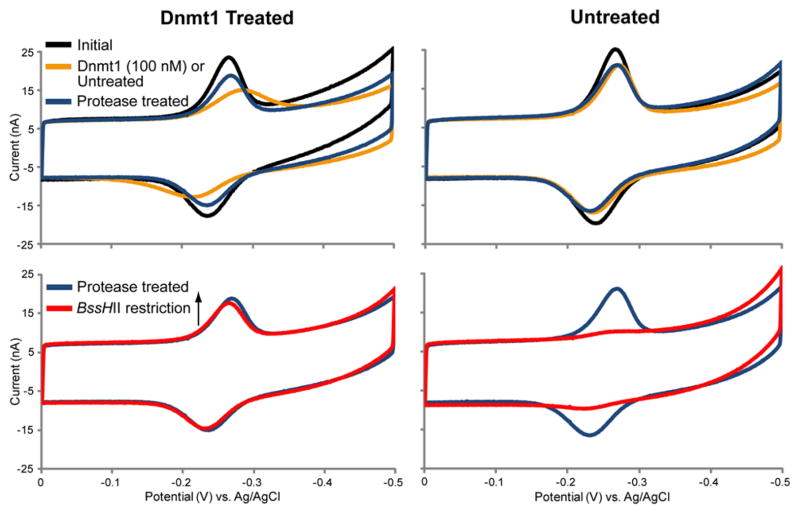
Figure 4.
Protease treatment restores peak sharpness for samples of high protein content. Chips were modified in all quadrants with the hemimethylated BssHII 22-mer. One half of the chip was treated with 100 nM Dnmt1 with 100 μg/mL BSA and 160 μM SAM (left column), while the other half of the chip was left untreated with 100 μg/mL BSA and 160 μM SAM alone (right column). Overlays of CV scans at various stages in the chip treatment are shown (top row) including the initial signal (black traces), signal after Dnmt1 treatment (orange traces), and signal after treatment with 1 μM protease (blue traces). The electrochemical signals used to quantify DNA methylation from these chips are also shown (bottom row) including the signal after protease treatment (blue traces, same as top row) and signal after BssHII restriction (red traces). All CV scans were performed in optimized scanning buffer (5 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 μM EDTA, 10% glycerol, pH 7) with an Ag/AgCl reference electrode at a 100 mV/s scan rate.
To address this issue, a protease treatment step was introduced to remove bound methyltransferases; following methyltransferase treatment, prior to BssHII treatment, chips were treated with a 1 μM solution of protease from Streptomyces griseus. Electrochemical scans of a chip with methyltransferase-treated and untreated quadrants at these sequential steps are shown in Figure 4. After the first 37°C heated incubation, signals from untreated electrodes show a small decrease in peak height, without peak broadening, that typically occurs the first time a chip is heated (77% ± 1% of the original peak height). In contrast, signals from electrodes treated with 100 nM Dnmt1 show a much larger decrease in peak height (46% ± 3% of the original peak height) with substantial peak broadening. After protease treatment, however, the relative peak heights of treated and untreated electrodes are equalized (74% ± 2% and 73% ± 1% of the original peak heights, respectively) and peak broadening of the Dnmt1-treated electrodes is reversed.
From this equal starting point, methylation can then be visualized by treatment with BssHII; signals from electrodes treated with 100 nM Dnmt1 are largely protected (89% ± 3%) while minimal signal remains for untreated electrodes (5% ± 1%). Although proteases are routinely used to remove interfering proteins in nucleic acid isolation and in situ hybridization (ISH) protocols, protease treatment has not yet been described as a strategy to enhance electrochemical biosensing on DNA-modified surfaces. Demonstrated here, protease treatment removes bound proteins, restores signal sharpness, and equalizes electrode signals such that Dnmt1 activity may be accurately quantified and compared over a wide range of methyltransferase concentrations.
Substrate Preference of Human Dnmt1
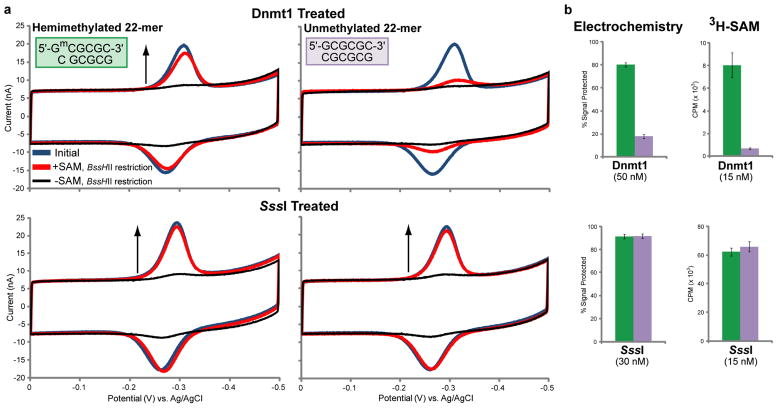
To measure the substrate preference and cofactor dependence of Dnmt1 electrochemically, multiplexed chips were modified in two quadrants with the hemimethylated BssHII 22-mer and in two quadrants with the unmethylated BssHII 22-mer. One quadrant of each DNA substrate was then treated with 50 nM Dnmt1 with SAM, while the other quadrant of each DNA substrate was treated with 50 nM Dnmt1 alone (Figure 5a, top row). As a comparison, chips were assembled and treated similarly with SssI (30 nM), which is a de novo methyltransferase and shows no preference for hemimethylated or unmethylated 5′-CG-3′ sites (Figure 5a, bottom row).44 For electrodes modified with the hemimethylated BssHII 22-mer and treated with Dnmt1 and SAM, substantial signal protection is observed (80% ± 2%). The same treatment shows minimal protection of the unmethylated BssHII 22-mer (18% ± 2%). In contrast, electrodes treated with 30 nM SssI and SAM show the same high level of signal protection regardless of whether the hemimethylated or unmethylated BssHII 22-mer is the substrate (91% ± 2% and 92% ± 2%, respectively). These quantified data are shown in Figure 5b, left column. For electrodes treated with Dnmt1 without SAM, a near complete lack of signal protection is observed for both the hemimethylated and unmethylated BssHII 22-mers (5% ± 1% and 4% ± 1%, respectively). Hemimethylated and unmethylated electrodes treated with SssI without SAM also show the same, minimal level of signal protection (5% ± 1% for both). These important, SAM-free controls further confirm that methylation is the mode of signal protection and demonstrate that both the hemimethylated and unmethylated BssHII 22-mers are cut equally by BssHII when not protected by methylation.
Figure 5.
Substrate preferences of SssI and Dnmt1 by electrochemical and 32H-SAM assays. (a) For electrochemistry, chips were modified on one half with the hemimethylated BssHII 22-mer (a, left column) and on the other half with the unmethylated BssHII 22-mer (a, right column). The BssHII 22-mer consists of the sequence 5′-HS- (CH2)6 – GACTGAGTACT GCGCGCACTGA-3′ with a methylene blue-modified complement. The hemimethylated (5′-GmCGCGC -3′) or unmethylated (5′-GCGCGC - 3′) BssHII restriction site is underlined. DNA protection was evaluated after treatment with 50 nM Dnmt1 (a, top row) or 30 nM SssI (a, bottom row), both with 160 μM SAM. For all plots, the initial signal (blue traces) and signal after BssHII (1,500 units/mL) treatment (red traces) are shown overlaid. Negative controls (treatment with Dnmt1 or SssI without SAM) are shown overlaid after BssHII treatment (black traces). (b) The cathodic peak areas of electrochemical data were quantified (b, left column) for hemimethylated (green bars) and unmethylated (lavender bars) DNA. A conventional 32H-SAM assay was also used to assess substrate preference (b, right column), using the same BssHII 22-mer sequence. Error bars for electrochemical data represent variation across 8 electrodes for each condition. Error bars for the 32H-SAM data represent the standard deviation across 3 replicates. All CV scans were performed in optimized scanning buffer (5 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 μM EDTA, 10% glycerol, pH 7) with an Ag/AgCl reference electrode at a 100 mV/s scan rate.
These electrochemical activity and substrate preference results for Dnmt1 and SssI were further validated by conventional 32H-SAM Assay. For these experiments, the same hemimethylated and unmethylated BssHII 22-mers, without redox probe or thiol modifications, were employed. Mirroring the electrochemical result, Dnmt1 shows substantially more activity on the hemimethylated BssHII 22-mer, while SssI shows the same activity on both the unmethylated and hemimethylated 22-mers (Figure 5b, right column).
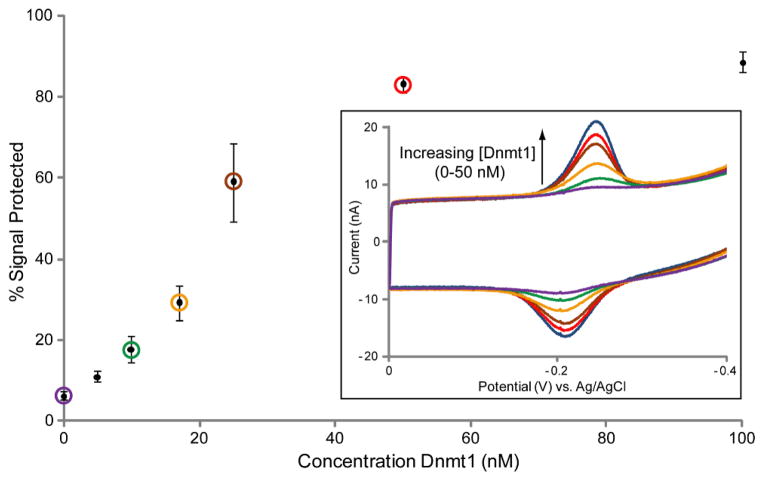
Concentration Dependence of Dnmt1 Methyltransferase Activity
With protease treatment to establish a uniform initial point of comparison, the concentration-dependent response of this assay for Dnmt1 activity was next evaluated. Multiplexed chips modified with the hemimethylated BssHII 22-mer were treated with a range of Dnmt1 concentrations (0 – 100 nM), followed by protease and BssHII treatments (Figure 6, inset). An activity curve was derived by compiling data from 4 chips with 4–12 electrodes measured at each concentration (Figure 6). Near complete signal protection is achieved by Dnmt1 concentrations of 50 nM and higher (83% ± 2% at 50 nM; 89% ± 3% at 100 nM). The dynamic range of Dnmt1 is much broader than that of SssI, spanning 5–50 nM. A detection limit of 5 nM Dnmt1 is the concentration below which signal protection is no longer distinguished from the signal that remains on untreated electrodes (11% ± 1% at 5 nM; 6% ± 1% for untreated electrodes).
Figure 6.
Concentration dependence of Dnmt1 Methyltransferase activity. Chips were modified in all quadrants with the hemimethylated BssHII 22-mer. DNA protection by various concentrations of Dnmt1 was evaluated side by side on the same chip, and CV scans from representative electrodes are shown overlaid (inset). Overlaid CV traces include the initial signal before BssHII treatment (blue), 50 nM Dnmt1 (red), 25 nM Dnmt1 (brown), 17 nM Dnmt1 (orange), 10 nM Dnmt1 (green), and untreated (purple). Quantification of the observed signal protection is displayed as a Dnmt1 activity curve where the circled points correspond to the concentrations represented by the overlaid CV traces. Error bars represent the standard deviation across 4–12 electrodes tested for each Dnmt1 concentration on 4 chips. All CV scans were performed in optimized scanning buffer (5 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 μM EDTA, 10% glycerol, pH 7) with an Ag/AgCl reference electrode at a 100 mV/s scan rate.
Comparison of Assay Responses by SssI and Dnmt1
Results from this electrochemical assay reflect the clear differences in activity between bacterial SssI and human Dnmt1, as well as their distinct roles as de novo and maintenance methyltransferases, respectively. Side by side electrochemical analysis of activity on unmethylated and hemimethylated DNA substrates shows the hemimethylated substrate preference of Dnmt1 and the absence of a preference for SssI, a result mirrored in 32H-SAM experiments. Additionally for this assay, while SssI exhibits a sharp, switch-like, signal protection response with a narrow dynamic range of 2–8 nM, the dynamic range for Dnmt1 activity spans a much broader 5–50 nM. For SssI, both the limit of detection and full dynamic range of the assay fall below its solution dissociation constant (Kd) of 11 nM.45 This indicates that methylation-induced signal protection is not limited by substrate access and affinity; once the concentration of SssI reaches a minimal level that allows appreciable binding, the DNA film is efficiently methylated and protected. In contrast, the Kd of 23 nM for Dnmt1 falls in the middle of the broad dynamic range measured by this assay.46
The contrasting dynamic ranges exhibited by SssI and Dnmt1 clearly reflect inherent differences in activity (as expected, parallel 32H-SAM experiments verify that the tested SssI is significantly more active than the tested Dnmt1), but may also reflect more subtle differences that are exaggerated by this surface platform. The broader dynamic range of Dnmt1 suggests that the DNA film is a substantially more difficult substrate matrix for Dnmt1 than it is for SssI. At nearly 4.5-fold larger than SssI, the size of Dnmt1 may be a limiting factor for methylation in the confined surface environment. Not only is the larger size likely a greater hindrance to turnover rates at the surface, Dnmt1 may also not be physically able to access all of the DNA sites in the film that SssI is able to access. The greater maximum signal-on magnitude achieved by SssI over Dnmt1 provides additional support that DNA access is more limited for Dnmt1. Notably, access to the DNA film is not a significant issue for the restriction enzymes used in this assay; not only are BstUI and BssHII comparable in size to SssI, but individual DNA duplexes are made more and more accessible as progressive restriction of the DNA film occurs.
We can take clues from these divergent responses by SssI and Dnmt1 to further refine the DNA-modified electrode platform that is used to carry out this assay. For assays that require binding of a target molecule to a nucleic acid probe on a surface, the physical accessibility of the nucleic acid probe is a critical factor that can dictate both the sensitivity and dynamic range of a biosensor platform.47 Notably, previous studies have shown that for self-assembled monolayers like those used in this work, thiolated DNA duplexes form films of heterogenenous density on gold surfaces.48 Thus for this assay, different DNA film morphologies (e.g. fully accessible duplexes spaced evenly across the surface or dense island patches of DNA for which only outer duplexes are accessible to protein activity), may present more or less challenging landscapes for a given methyltransferase to access and fully protect from restriction. The film density and morphology of DNA-modified electrodes must be a central point of consideration for modulating the sensitivity and dynamic range of this assay for different applications.
Comparison to Current Approaches for the Detection of Methyltransferase Activity
As compared to other approaches in the literature to measure methyltransferase activity, a notable advantage of this assay is the small sample volume (10 μL) required for analysis. For colorimetric and fluorescence methods in solution, sample volumes typically range from at least 20 – 150 μL.21–24,49,50 Furthermore, the sample volume of our assay is only restricted by our current electrochemical platform design and thus hardly at a lower limit. Unlike other assays that are constrained by the configuration and sensitivity of spectrophotometers and fluorescence readers, the flexibility of electrochemical platforms inherently supports miniaturization. Still, with our current configuration, SssI activity can be observed by as little as 20 fmoles (0.8 ng) of protein while Dnmt1 activity can be observed by as little as 50 fmoles (9 ng) of protein.
Several electrochemical assays have been described for the detection of SssI activity based on methylation protection of DNA films from cutting by the HpaII restriction enzyme.32–35 Our assay advances beyond these previous electrochemical approaches in that it allows measurements on both hemimethylated and unmethylated DNA substrates. Thus, our assay uniquely permits the detection of human methyltransferase activity by an electrochemical method.
Currently, commercially available ELISA-like kits are the main alternative to radioactive methods for the detection of human methyltransferase activity in both purified samples and crude cell lysates. The sensitivity and dynamic range of our electrochemical assay for Dnmt1 detection is comparable to these commercially available kits,49,50 but the cost and complexity of necessary reagents and equipment for our approach is substantially less. These commercial assays require primary and secondary antibodies along with colorimetric reagents or fluorescent labels and an absorbance or fluorescence microplate reader.49,50 In contrast for our assay, multiplexed chips and DNA substrates may be inexpensively fabricated in bulk and the BssHII restriction enzyme is available cheaply from commercial sources. Furthermore, the advantages of electrochemical instrumentation for biosensing are well established; potentiostats are relatively simple, inexpensive devices that require minimal maintenance and can be made portable, thereby making them accessible to a wide range of research and clinical settings.25,26 Beyond cost and ease of implementation, an additional advantage of our assay is that the activity of a sample on hemimethylated and unmethylated DNA substrates may be measured separately and compared on the same device. This capacity to assess the maintenance vs. de novo methyltransferase activity of a sample is critical for studies involving human methyltransferases because of the profound impact of these specific roles on cancer processes.
CONCLUSIONS
Described here is a multiplexed, signal-on, electrochemical assay for the sensitive detection of bacterial and human methyltransferase activity. This non-radioactive, antibody-free assay is robust and generally requires less sample volume than other methods. The electrochemical basis of this assay provides numerous advantages, including minimal, portable equipment, inexpensive reagents, and a format that may be easily adapted to even more effective electrode architectures. The unique capability of this assay to provide a side-by-side report of activity on unmethylated and hemimethylated DNA substrates is a key feature for studies with human methyltransferases. As we continue to discover the epigenetic basis of cancerous transformation, the demand for effective and inexpensive strategies to detect human methyltransferase activity will continue to grow. Widely accessible approaches, like the electrochemical assay presented here, will be necessary to meet this demand for cancer detection and treatment.
Acknowledgments
This work was supported by NIH GM61077. We also thank the Parsons Foundation for a fellowship to NBM. We thank C. Pheeney and P. Bartels for assistance with multiplexed chip fabrication. Multiplexed chip fabrication was completed in the Kavli Nanoscience Institute at Caltech.
References
- 1.Jeltsch A. ChemBioChem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Miranda TB. J Cell Phys. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 3.Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R, Akbarian S, Tudor M, Jaenisch R. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 5.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 7.Das PM, Singal R. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Herman JG. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 9.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 10.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. Nuc Acid Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajendran G, Shanmuganandam K, Bendre A, Mujumdar D, Goel A, Shiras A. J Neurooncol. 2011;104:483–494. doi: 10.1007/s11060-010-0520-2. [DOI] [PubMed] [Google Scholar]
- 12.Shukla V, Coumoul X, Lahusen T, Wang R, Xu X, Vassilopoulos A, Xiao C, Lee M, Man Y, Ouchi M, Ouchi T, Deng C. Cell Res. 2010;20:1201–1215. doi: 10.1038/cr.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roll JD, Rivenbark AG, Jones WD, Coleman WB. Mol Cancer. 2008;7:15–29. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pradhan S, Bacolla A, Wells RD, Roberts RJ. J Biol Chem. 1995;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 15.Okano M, Bell DW, Haber DA, Li E. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 16.Issa J. Nat Rev. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 17.Jurkowska RZ, Ceccaldi A, Zhang Y, Ariimondo PB, Jeltsch A. Methods Mol Biol. 2011;791:157–177. doi: 10.1007/978-1-61779-316-5_13. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Rohde C, Tierling S, Stamerjohanns H, Reinhardt R, Walter J, Jeltsch A. Methods Mol Biol. 2009;507:177–187. doi: 10.1007/978-1-59745-522-0_14. [DOI] [PubMed] [Google Scholar]
- 19.Friso S, Choi S, Dolnikowski GG, Selhub J. Anal Chem. 2002;74:4526–4531. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 20.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. Anal Biochem. 2006;15:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Graves TL, Zhang Y, Scott JE. Anal Biochem. 2008;373:296–306. doi: 10.1016/j.ab.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strivers JT, Ye Y. Anal Biochem. 2010;401:168–172. doi: 10.1016/j.ab.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood RJ, McKelvie JC, Maynard-Smith MD, Roach PL. Nuc Acids Res. 2010;38:1–11. doi: 10.1093/nar/gkq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Chen F, Wu Y, Dong Y, Fan C. Biosens Bioelect. 2013;42:56–61. doi: 10.1016/j.bios.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Sadik OA, Aluoch AO, Zhou A. Biosens Bioelect. 2009;24:2749–2765. doi: 10.1016/j.bios.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Mai Z, Dai Z, Zou X. Chem Commun. 2010;46:7781–7783. doi: 10.1039/c0cc00983k. [DOI] [PubMed] [Google Scholar]
- 28.He X, Su J, Wang Y, Wang K, Ni X, Chen Z. Biosens Bioelectron. 2011;28:298–303. doi: 10.1016/j.bios.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, He X, Wang K, Su J, Chen Z, Yan G, Du Y. Biosens Bioelectron. 2013;41:238–243. doi: 10.1016/j.bios.2012.08.034. [DOI] [PubMed] [Google Scholar]
- 30.Su J, He X, Wang Y, Wang K, Chen Z, Yan G. Biosens Bioelectron. 2012;36:123–128. doi: 10.1016/j.bios.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Liu S, Jiang J, Shen G, Yu R. Chem Commun. 2012;48:6280–6282. doi: 10.1039/c2cc32397d. [DOI] [PubMed] [Google Scholar]
- 32.Liu S, Wu P, Li W, Zhang H, Cai C. Chem Commun. 2011;47:2844–2846. doi: 10.1039/c0cc05153e. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Wu P, Zhang H, Cai C. Anal Chem. 2012;84:7583–7590. doi: 10.1021/ac301990f. [DOI] [PubMed] [Google Scholar]
- 34.Yin H, Zhou Y, Xu Z, Chen L, Zhang D, Ai S. Biosens Bioelectron. 2012;41:492–497. doi: 10.1016/j.bios.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Wang M, Xu Z, Chen L, Yin H, Ai S. Anal Chem. 2012;84:9072–9078. doi: 10.1021/ac301620m. [DOI] [PubMed] [Google Scholar]
- 36.Yamada H, Tanabe K, Nishimoto S. Org Biomol Chem. 2008;6:272–277. doi: 10.1039/b715260d. [DOI] [PubMed] [Google Scholar]
- 37.Slinker JD, Muren NB, Gorodetsky AA, Barton JK. J Am Chem Soc. 2010;132:2769–2774. doi: 10.1021/ja909915m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boon EM, Salas JE, Barton JK. Nat Biotechnol. 2002;20:282–286. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 39.Slinker JD, Muren NB, Renfrew SE, Barton JK. Nat Chem. 2011;3:230–235. doi: 10.1038/nchem.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pheeney CG, Barton JK. Langmuir. 2012;28:7063–7070. doi: 10.1021/la300566x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams RLP, McKay EL, Craig LM, Burdon RH. Biochim Biophys Acta. 1979;561:345–357. doi: 10.1016/0005-2787(79)90143-6. [DOI] [PubMed] [Google Scholar]
- 42.Renbaum P, Abrahamove D, Fainsod A, Wilson GG, Rottem S, Razin A. Nuc Acids Res. 1990;18:1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renbaum P, Razin A. J Mol Biol. 1995;248:19–26. doi: 10.1006/jmbi.1995.0199. [DOI] [PubMed] [Google Scholar]
- 44.Flynn J, Glickman JF, Reich NO. Biochemistry. 1996;35:7308–7315. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- 45.Darii MV, Kirsanova OV, Drutsa VL, Kochetkov SN, Gromova ES. Mol Biol. 2007;41:110–117. [PubMed] [Google Scholar]
- 46.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. J Biol Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soleymani L, Fang Z, Sargent EH, Kelley SO. Nat Nanotech. 2009;4:844–848. doi: 10.1038/nnano.2009.276. [DOI] [PubMed] [Google Scholar]
- 48.Murphy JN, Cheng AKH, Yu H, Bizzotto D. J Am Chem Soc. 2009;131:4042–4050. doi: 10.1021/ja808696p. [DOI] [PubMed] [Google Scholar]
- 49.BPS Bioscience Inc. DNMT Universal Assay Kit. 2013 < http://www.bpsbioscience.com/methyltransferase/assay-kit/dnmt-universal-assay-kit-52035>.
- 50.Epigentek Group Inc. EpiQuik DNA Methyltransferase Activity/Inhibition Assay Kit. 2013 < http://www.epigentek.com/catalog/epiquik-dna-methyltransferase-dnmt-activityinhibition-assay-kit-p-98.html>.