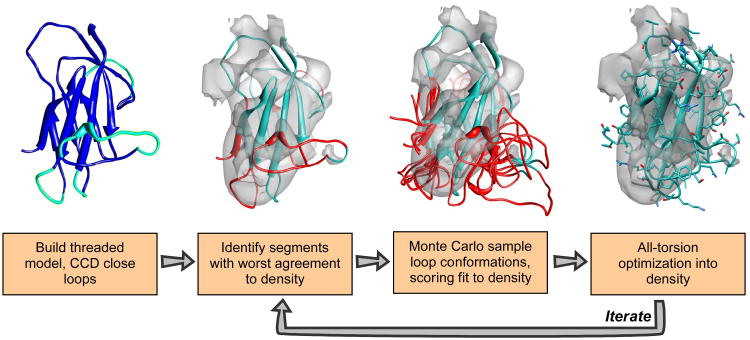
Figure 1.
The comparative modeling into density protocol. We initially build a threaded model from some alignment (blue), using cyclic coordinate descent to close gaps in the alignment (cyan). We then dock this threaded model into density, and identify regions that have a poor local agreement with the density data (red). We aggressively resample the conformations in these regions, scoring each potential conformation with Rosetta's low-resolution energy function together with an agreement-to-density score. Finally, we optimize sidechain rotamers and minimize all backbone and sidechain torsions using Rosetta's high-resolution potential, also augmented with this agreement-to-density score. We iterate over these final three steps until the lowest-energy models converge, at each iteration enriching our population for those models with both favorable Rosetta energy as well as good fit to density.