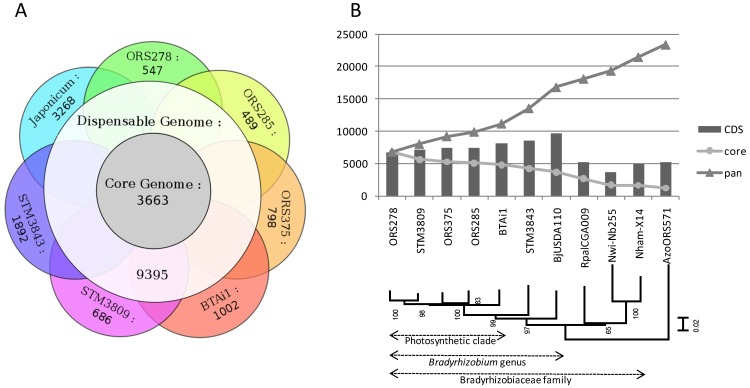
Figure 2.
(A) Comparative gene orthology between seven genomes of Bradyrhizobium. Numbers indicates the shared gene sets between groups of strains, satisfying the following criteria: BBH (bidirectional best hits) with 40% amino-acid identity on 80% of the smallest protein; (B) Core and pangenome gene count in the Bradyrhizobiaceae family. CDS indicates the number of protein-coding genes in each genome listed in x-axis. The y-axis indicates the number of genes in core and pangenome when adding genomes in the x-axis to the comparative analysis. The presented tree was built by Maximum likelihood (GTR + I + G model) on a partition of 5 taxonomic marker genes (atpD, dnaK, recA, rpoB, rpoD), with bootstrapped nodes (100 replicates). Japonicum/Bj: B. japonicum USDA110; Rpal: Rhodopseudomonas palustris; Nwi: Nitrobacter winogradskii; Nham: Nitrobacter hamburgensis; Azo: Azorhizobium caulinodans.