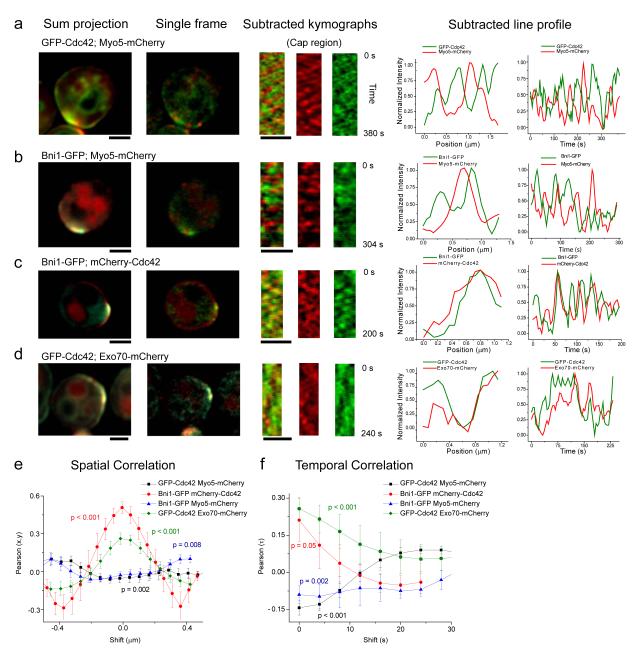
Figure 1. Spatial organization of endocytic and exocytic domains relative to Cdc42 distribution.
a-d. Left most column shows representative time-summed images of the localization of the indicated proteins. These images show the overall presence of the analyzed proteins in the polar cap. The column second from the left shows example single time point images showing punctate appearance of the proteins observed. The three columns that follow show sum-subtracted kymographs (merged followed by mCherry and GFP) of the polar cap region from image series showing anti-correlated (a,b) or correlated (c,d) fluctuations of the two proteins in each pair as indicated. Raw (non-average subtracted) kymographs are shown in Supplementary Fig. S1. The second from right plot shows an example fluorescence profile at a single time point from the kymographs to the left, whereas the right most graph shows a time trace at a single cortical location. Scale bars: 2 μm.
e,f. Average Pearson cross-correlation between red and green fluorescence spatially (along rows of the kymographs) (e) and temporally (columns of kymographs) (f). Negative correlation was observed for Bni1 with Myo5, and Cdc42 with Myo5, whereas positive correlation was observed between Bni1 and Cdc42 and Exo70 and Cdc42 at small space (e) or time shifts (f). Plots show mean and standard error of the mean (SEM) from 6 to 8 kymographs per pair. P-values are based on a one-component student’s t-test, compared to zero, or a random distribution.