Fig. 1.
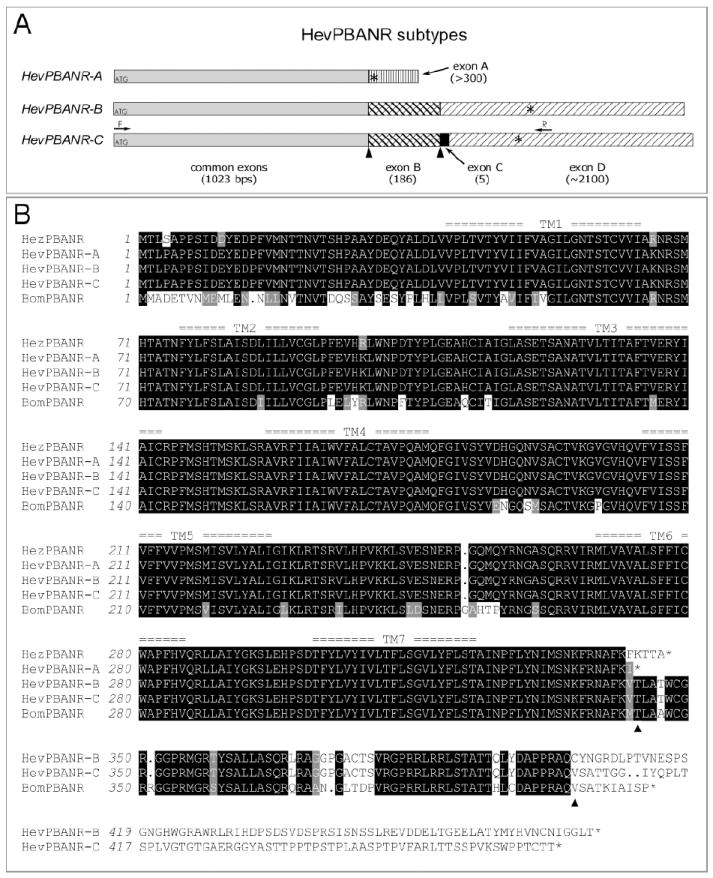
Three subtypes of HevPBANR.
(A) Schematic diagram showing the organization of putative exons in three HevPBANR subtypes. cDNA sequence comparisons between subtypes suggest the cDNA of each receptor subtype consists of a common region (gray bar) and four additional exons (variously patterned bars; exon A, -B, -C and -D), which are arranged in subtype-specific configurations. Numbers in parentheses indicate exon lengths. Translation initiation and termination sites are indicated by ATGs and asterisks (*) respectively. Arrows labeled with either F or R indicate primer binding sites used to amplify the entire open reading frame of HevPBANR-C from pheromone glands. (B) Protein sequence alignment of three HevPBANR subtypes described in this study. Helicoverpa zea PBANR (HezPBANR, GenBank accession number, AY319852) and Bombyx mori PBANR (BomPBANR, AB181298). Note that C-terminal region of HevPBANR subtype-A is quite similar to that of HezPBANR, while subtypes-B, and C are similar to BomPBANR. Triangles indicate junctions between exons corresponding to those labeled with triangles in (A). Seven transmembrane domains are numbered as TM1- TM7. Identical amino acids are highlighted in black shadow and conservative amino acids are in light gray. Dots are gaps introduced for alignment.
