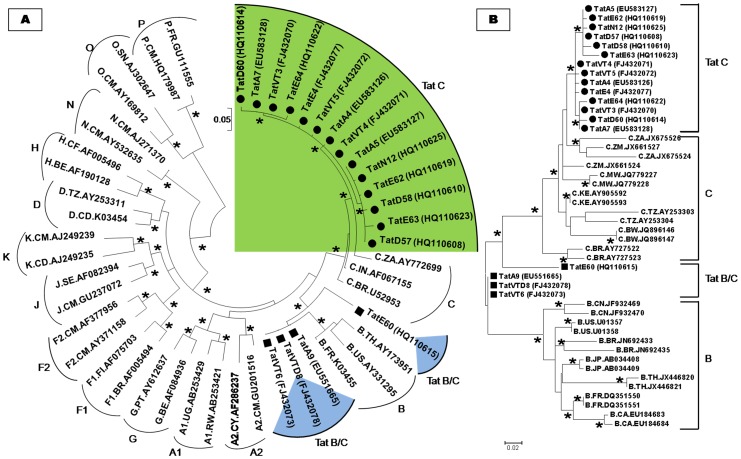
Figure 1. HIV-1 sub-typing based on global subtype references.
A) The phylogenetic tree of Tat exon-1 variants with M (A to K including A1, A2, F1, and F2), N, O and P groups. B) The phylogenetic tree of Tat exon-1 variants with global subtype B from countries such as China (CN), France (FR), Japan (JP), Thailand (TH), United States (US), Canada (CA) and Brazil (BR) and global subtype C from countries such as Botswana (BW), Tanzania (TZ), South Africa (ZA), Zambia (ZM), Kenya (KE), Malawi (MW) and Brazil (BR) sequences. Each reference sequence was labelled with a subtype, followed by country of isolation and accession number. Filled circles represent our C variants and filled rectangles represent our B/C recombinants. The bootstrap probability (>65%, 1,000 replicates) was showed with asterisk (*) at the corresponding nodes of the tree and the scale bar represents the selection distance of 0.05 and 0.02 nucleotides per position in the sequence.