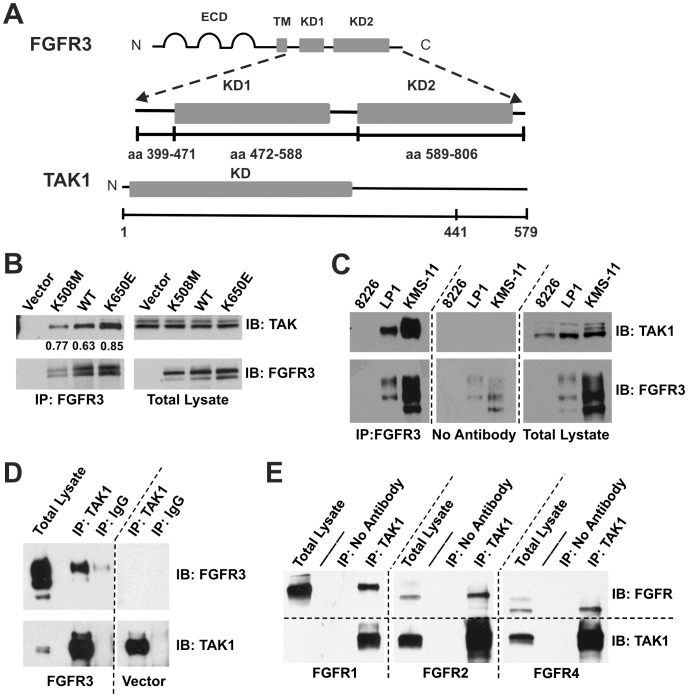
Figure 1. TAK1 interacts with FGFR3.
(A) Schematic of FGFR3 and TAK1 domains used for yeast two-hybrid screening and subsequent mapping of the interaction in yeast. (B) Endogenous TAK1 interacts with kinase-dead (K508M), wild-type, and constitutively active (K650E) FGFR3 in HeLa cells. Numerical values represent the ratio of TAK1 co-precipitated with FGFR3. (C) FGFR3 and TAK1 (both endogenous) interact in LP1 (FGFR3WT) and KMS-11 (FGFR3Y373C) multiple myeloma cell lines. The 8226 line is negative for FGFR3. (D) Endogenous TAK1 interacts with FGFR3 in MGHU3 bladder cancer cells transfected with wild-type FGFR3. MGHU3 also express the FGFR3 activating mutation, Y375C. (E) TAK1 (endogenous) interacts with overexpressed FGFR1, −2, and −4 in HEK293 cells. TAK1 in the FGFR1-transfected total lysate is detectable upon longer exposure (data not shown). For all blots (B-E), immunoprecipitations were performed from 1mg total lysate using the antibody indicated. Blots were first probed for the interaction partner being tested, then stripped and re-probed for the immunoprecipitated protein. 20 µg total lysate was similarly probed to control for expression and loading. Arrow indicates TAK1. Multiple FGFR3 bands represent various glycosylation intermediates and appear as previously published [45], [85]. Four independent experiments were performed for each panel.