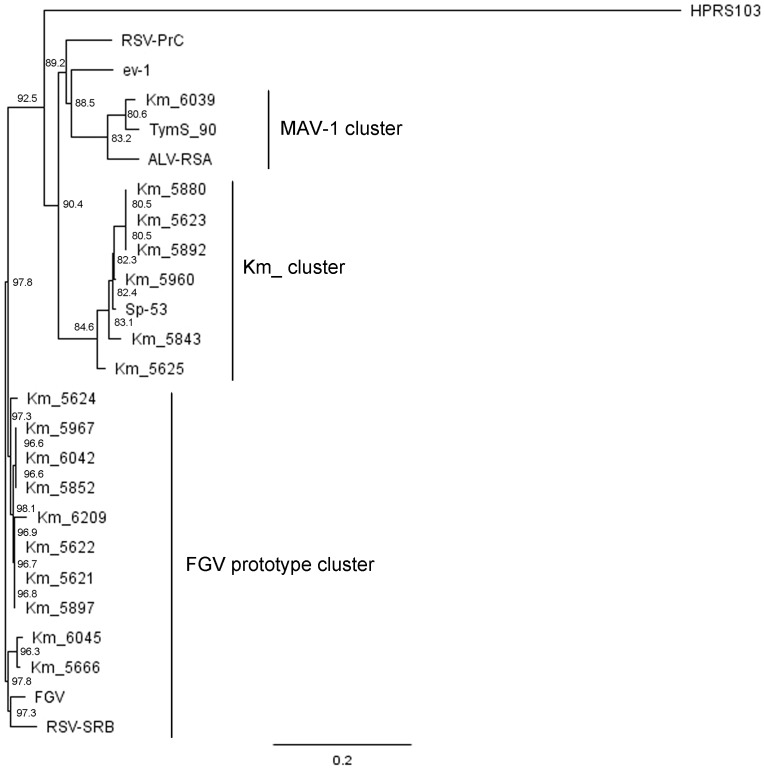
Figure 2. Phylogenic tree based on envSU region of the isolates.
Phylogenic tree constructed by the neighbor-joining method showing the relationships among the isolates, FGV, Sp-53 (FGV variant) and standard ALSVs based on the SU region of the env gene. The amino acid sequences were aligned with the Clustal W 1.8.3 program. Bootstrap values of 1000 trials using the neighbor-joining method. Scale bar corresponds to a distance of 0.2 as the frequency of amino acid substitutions in the pairwise comparison of two sequences according to the Kimura two-parameter method.