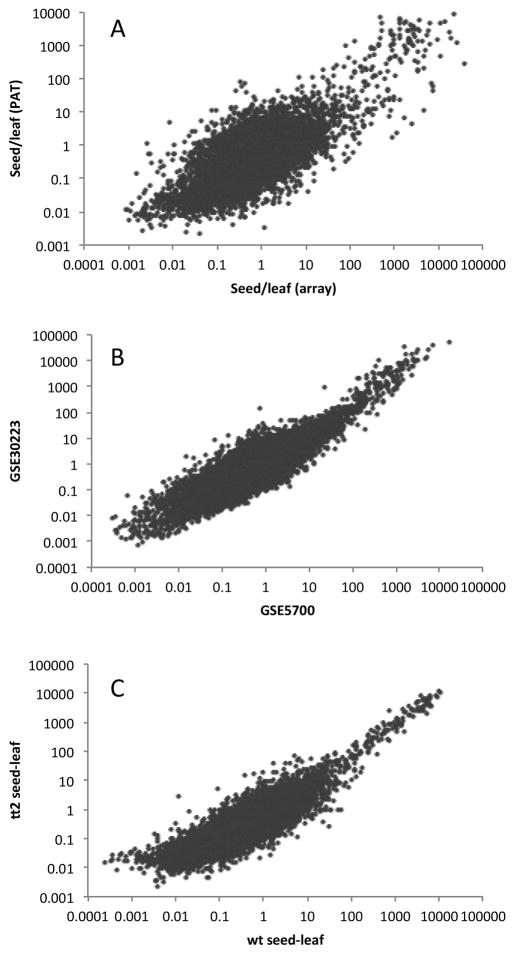
Figure 5.
Comparison of seed/leaf gene expression ratios obtained using PATs and microarrays. A. Plot of values (displayed using a log10 scale) obtained using 8 microarray datasets (five seed and three leaf) and 13 PAT datasets (five seed and eight leaf); the PAT datasets were generated from tags prepared using Methods B1 and B2. The Pearson correlation coefficient for the linear relationship of the log-log plot was 0.73. B. Plot of values (displayed using a log10 scale) obtained using either the GSE30223 or GSE5700 array data as sources of seed gene expression (in both cases, GSE5630 was the source for leaf gene expression). The Pearson correlation coefficient for the linear relationship of the log-log plot was 0.89. C. Plot of values (displayed using a log10 scale) obtained using either the tt2 or wt seed PATs (prepared using Methods B2 and B1, respectively) as sources of seed gene expression (in both cases, all 8 leaf PAT preparations made using Methods B1 and B2 were the source for leaf gene expression). The Pearson correlation coefficient for the linear relationship of the log-log plot was 0.87. For all three plots, 9661 genes were analyzed (this was the number that had average expression values greater than 10 tpm in either the leaf or seed samples).