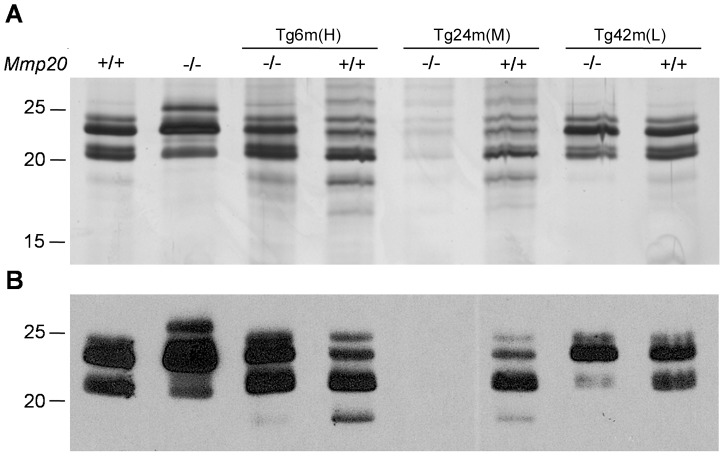
Figure 8. Enamel matrix protein profile and immunoblotting of amelogenin splice and cleavage products.
Total extracted enamel matrix proteins (mostly amelogenins) from 5-day old molars were run on an SDS PAGE gel (A). Comparison of the first two lanes containing extracted wild-type or Mmp20 null enamel matrix proteins demonstrate that the null enamel has a prominent band at approximately 27 kDa that is only weakly present in the wild-type lane indicating that this band is cleaved by MMP20. Also a doublet just above 20 kDa was present in the wild-type lane but was a single band in the null lane indicating that one of the wild-type bands is an MMP20 cleavage product. The protein profile for the Tg6m (H) transgene in the Mmp20−/− background was similar to that of the protein extracted from wild-type enamel. However, the protein profile for Tg6m (H) and Tg24m (M) in the Mmp20+/+ background had a strong band below the 20 kDa marker and a weaker band beneath that indicating that more cleavage products than normal were present. Both transgenes in the wild-type background had bands above 20 kDa that were less prominent than the bands observed from wild-type enamel with no transgene. In contrast, regardless of the Mmp20 background, the Tg42m (L) positive mice had protein banding patterns that looked substantially like the wild-type results. Amelogenin immunoblot results from the various geneotypes (B) were similar to the protein gel results. An approximate 27-kDa amelogenin band was present in the Mmp20 null but not wild-type lanes. In the wild-type background, both Tg6m (H) and Tg24m (M) transgenes had prominent amelogenin bands that located below 20 kDa and the bands above 20 kDa appeared less prominent than those observed in the Tg– wild-type lane. Also the amelogenin profile of Tg42m (L) in the wild-type background looked similar to that of wild-type mice.