Abstract
Indigenous Culicoides biting midges are suggested to be putative vectors for the recently emerged Schmallenberg virus (SBV) based on SBV RNA detection in field-caught midges. Furthermore, SBV replication and dissemination has been evidenced in C. sonorensis under laboratory conditions. After SBV had been detected in Culicoides biting midges from Belgium in August 2011, it spread all over the country by the end of 2011, as evidenced by very high between-herd seroprevalence rates in sheep and cattle. This study investigated if a renewed SBV circulation in midges occurred in 2012 in the context of high seroprevalence in the animal host population and evaluated if a recently proposed realtime RT-PCR approach that is meant to allow assessing the vector competence of Culicoides for SBV and bluetongue virus under laboratory conditions was applicable to field-caught midges. Therefore midges caught with 12 OVI traps in four different regions in Belgium between May and November 2012, were morphologically identified, age graded, pooled and tested for the presence of SBV RNA by realtime RT-PCR. The results demonstrate that although no SBV could be detected in nulliparous midges caught in May 2012, a renewed but short lived circulation of SBV in parous midges belonging to the subgenus Avaritia occured in August 2012 at all four regions. The infection prevalence reached up to 2.86% in the south of Belgium, the region where a lower seroprevalence was found at the end of 2011 than in the rest of the country. Furthermore, a frequency analysis of the Ct values obtained for 31 SBV-S segment positive pools of Avaritia midges showed a clear bimodal distribution with peaks of Ct values between 21–24 and 33–36. This closely resembles the laboratory results obtained for SBV infection of C. sonorensis and implicates indigenous midges belonging to the subgenus Avaritia as competent vectors for SBV.
Introduction
Schmallenberg virus is an orthobunyavirus belonging to the Simbu serogroup and was first identified in 2011 [1]. It causes a non-specific syndrome including high fever, decrease in milk production and severe diarrhoea in adult cattle [1] and is furthermore responsible for abortions, stillbirths and congenital malformations such as the hydranencephaly-arthrogryposis syndrome in cattle, sheep and goat [2]–[4]. Since its initial appearance in Germany and The Netherlands, followed by Belgium, the United Kingdom and France, SBV has now spread over much of Europe and beyond [5]–[8].
Culicoides biting midges have in the meantime been implicated as putative vectors of SBV what is in line with the knowledge that related viruses belonging to the same serogroup like Akabane and Aino virus are transmitted by midges and mosquitoes [9]–[12]. Several studies have detected SBV in (heads of) field caught Obsoletus complex, C. dewulfi, C. chiopterus and C. punctatus midges [13]–[19] and SBV replication and dissemination in C. sonorensis midges has been shown under laboratory conditions [20].
In Belgium, SBV had spread all over the country by the end of the vector season of 2011 as evidenced by a very high between-herd seroprevalence in sheep and goats [21], [22]. In the southern part of the country, a relatively lower within-herd seroprevalence was found. This was in line with the observation that SBV positive Culicoides could be found at different sampling regions in the country, except in the most southern trapping locations [13] (unpublished results).
Despite the overall presence of anti-SBV antibodies in the host populations [21]–[23] which have been shown to protect against challenge infections under experimental conditions [24], [25], clear indications of a renewed SBV circulation in 2012 in sheep and cattle in Belgium and surrounding countries have been found [26]–[29]. In this study, the presence of SBV in field trapped Culicoides at different locations in Belgium in 2012 was examined and compared with the results from midges in 2011. Furthermore the results were used to evaluate a recently proposed method to assess vector competence of Culicoides for arboviruses via semi-quantitative RT-PCR [20], [30].
Materials and Methods
Culicoides Trapping
From May till November 2012, Culicoides were caught with Onderstepoort Veterinary Institute (OVI) traps [31] at 12 different locations (Table 1) covering 4 different regions of Belgium (Figure 1): Antwerp (north-east), Liège (east), Gembloux (centre) and Libramont (south). During that time period, Culicoides were caught biweekly during one night with the black light traps. Attracted insects were caught in a container containing 60% ethanol. All OVI traps were installed at places where livestock was present in the immediate vicinity (Table 1).
Table 1. Detailed information on Culicoides trapping locations.
| location | livestock | longitude (°E) | latitute (°N) |
| Antwerp | |||
| Betekom | sheep, deer, chickens | 4.79206 | 51.00200 |
| Varendonk | cows | 4.954160 | 51.085820 |
| Berlaar | cows | 4.665713 | 51.118610 |
| Nijlen | cows | 4.693747 | 51.159744 |
| Viersel | cows | 4.63627 | 51.18454 |
| Liège | |||
| Amay | Chickens, rabbits, sheep | 5.185050 | 50.33694 |
| Boncelles | horses | 5.554803 | 50.567739 |
| Sart-Tilman | cows, sheep, goats,chickens, horses | 5.587336 | 50.576544 |
| Nandrin | horses, cows, pigs | 5.358419 | 50.528464 |
| Gembloux | cows, sheep, pigs | 4.72662 | 50.56509 |
| Libramont | sheep | 5.35956 | 49.92881 |
| cows | 5.35636 | 49.92931 |
Figure 1. Culicoides trapping locations during the Culicoides monitoring of 2012.
Ethics Statement
A permission of access and realization of light trapping of Culicoides was given by the farmers of the different sampling locations. No protected species were sampled during this study.
Morphological Identification, Determination of the Physiological Status and Pool Preparation
The biting midges of each capture place and time were kept separately and were morphologically identified under the stereomicroscope using the key of Delécolle [32] and further stored in 80% ethanol. Culicoides from Antwerp, Gembloux and Libramont were identified at subgenus level and female midges belonging to the following subgenera were further examined: Avaritia (C. obsoletus, C. scoticus, C. dewulfi, C. chiopterus), Culicoides (C.pulicaris, C. punctatus, C. impunctatus, C. lupicaris, C. newsteadi, C. deltus, C. grisescens, C. fagineus) and Monoculicoides (C. nubeculosus, C. riethi, C. puncticollis). Midges belonging to other subgenera were grouped in separate pools designated as ‘other species’. Culicoides caught in the region of Liège were identified at species or complex level and females of the following species or complexes were further examined: Obsoletus complex, C. dewulfi, C. chiopterus, C. pulicaris and C. furcillatus. Before preparing subgenus, complex or species specific pools of maximum 20 whole Culicoides, their physiological status was determined as ‘nulliparous’, ‘parous non-engorged’, ‘blood present’ or ‘blood and eggs present’ as described by Fassotte et al. (2008) [33]. Sixty nine pools were prepared representing 1,359 nulliparous females caught in May in the region of Antwerp (Betekom, Nijlen, Varendonk) and Gembloux, while all other pools only contained parous non-engorged females (904 pools representing 17,461 midges).
rRT-PCR Analysis of Pools of Culicoides
To examine the presence of SBV in Culicoides, pools were analyzed by real-time reverse transcription PCR (rRT-PCR) as described before [13]. Briefly, each pool was homogenized in 500 µl Trizol (Life Technologies, Ghent, Belgium) with a 5 mm steel bead (Qiagen, Hilden, Germany) by high speed shaking (3 min, 25 Hz) in a TissueLyser (Qiagen). Following the manufacturer’s instruction, total RNA was extracted from the aqueous phase using the MagMAX Total Nucleic Acid Isolation kit and the MagMAX Express-24 purification system (Life Technologies). RNA was eluted in 90 µl elution buffer. The presence of SBV RNA was analyzed by using the AgPath-ID One Step RT-PCR kit (Life Technologies) following the manufacturer’s instructions in a duplex rRT-PCR for detection of the SBV-S segment [34] and the 18S rRNA from Culicoides [35] as an internal control for RNA extraction and amplification. Pools positive for the SBV-S segment were subjected to another rRT-PCR detecting the L segment of the virus [13] (primers and probe sequences were kindly provided by FLI, Germany) using the same one step RT-PCR kit for confirmation. When this second rRT-PCR resulted in data difficult to interpret due to Ct values derived from atypical fluorescence amplification curves, the RNA extract was retested with the same primers but in a two-step PCR with the FastStart TaqMan Probe Master kit (Roche, Basel, Switzerland) following the manufacturer’s instructions after reverse transcription using the M-MLV reverse transcriptase system (Life Technologies, Ghent, Belgium). In the end, only pools that were positive for the S and L segment were considered as SBV positive.
Results and Discussion
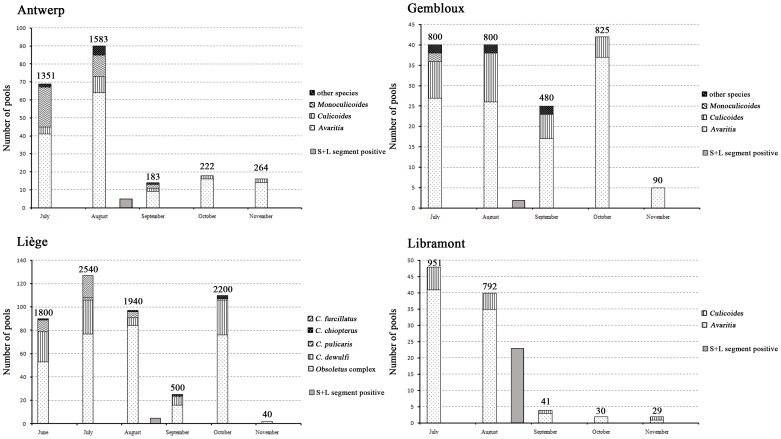
The extensive spread of SBV in 2011 in Belgium associated with the induction of high between- and within-herd seroprevalence rates in cattle and sheep [21], [22] raised the question at the beginning of the vector season 2012 if a renewed SBV circulation in hosts and Culicoides would be observed. The results of this study show that in all 4 regions where Culicoides were sampled, SBV positive pools were found at the beginning of August 2012 (Figure 2), confirming the presence of SBV. Despite the examination of a similar number of pools containing midges caught in July, no SBV positive Culicoides could be detected then. These results correlate well with a report describing that SBV was found in a single sheep belonging to a flock held in Namur (Belgium) around mid-July 2012, while most other sheep of the flock became viremic only between mid-August and late September [28]. A limited number of reported SBV positive newborn calves at the beginning of 2013 [27] and a few SBV positive aborted lambs and calves diagnosed at the Belgian reference laboratory CODA-CERVA from November 2012 onwards support an SBV circulation during August 2012.
Figure 2. Overview of pools of Culicoides examined for the presence of Schmallenberg virus originating from 4 trapping regions in 2012.
The numbers mentioned above the bars indicate the exact number of Culicoides tested.
At Libramont in the south of Belgium, the observed number of positive pools in 2012 was unexpectedly high with 57% (20/35) of Avaritia pools and 60% (3/5) of Culicoides pools being rRT-PCR positive (Figure 2, Table 2). If only 1 midge per pool was SBV positive, this would corresponds to an infection rate in August 2012 at Libramont of 2.86 and 3.26% in the subgenera Avaritia and Culicoides, respectively. These percentages are similar to the infection rates of 3.6 and 2.4% reported in Obsoletus complex midges in September 2011 in Antwerp and October 2011 in Liège respectively [13] (unpublished results). The infection rates of 2011 were however reached in the setting of a naïve host population while most potential host individuals were considered seropositive in 2012. It should be taken into account that in the 2011 study, examined pools contained only heads whilst in the present study whole midges were used. Since Ct values for SBV tend to be higher in heads of infected midges than in their abdomen [20], it can be assumed that the true infection rate in 2011 was possibly even higher than the reported values. The most probable explanation for the high infection rate in midges in the south of Belgium is the presence of hosts that had not been infected during the SBV epidemic in 2011. This hypothesis is supported by the reported relatively lower seroprevalence in sheep and cattle in the south of Belgium at the end of 2011 [21], [22] and the presence of a partially seronegative population of wild cervids in southern Belgium at that time [23]. If a fast drop of the acquired immune protection or a non-protective immune response induced by the previous infection would be considered as alternative explanations, it would seem logical that in the other examined regions infection prevalences should be similar to Libramont.
Table 2. Detailed overview of Schmallenberg virus positive pools of Culicoides.
| Ct value | |||||
| location | subgenus/species | collection date | I.C. | S segment | L segment |
| Antwerp | |||||
| Betekom | Avaritia | 01 Aug 2012 | 17.07 | 24.73 | 26.69 |
| Berlaar | Avaritia | 03 Aug 2012 | 13.05 | 22.01 | 23.82 |
| Berlaar | Avaritia | 03 Aug 2012 | 14.25 | 25 | 26.13 |
| Viersel | Avaritia | 14 Aug 2012 | 9.84 | 28.09 | 31.89 |
| Berlaar | Avaritia | 14 Aug 2012 | 10.87 | 23.67 | 26.32 |
| Liège | |||||
| Nandrin | Obsoletus complex | 09 Aug 2012 | 11.6 | 27.8 | 30.06 |
| Nandrin | Obsoletus complex | 09 Aug 2012 | 11.3 | 34.09 | 35.18 |
| Boncelles | Obsoletus complex | 09 Aug 2012 | 11.02 | 30.34 | 32.38 |
| Amay | Obsoletus complex | 22 Aug 2012 | 11.64 | 21.6 | 23.53 |
| Nandrin | Obsoletus complex | 22 Aug 2012 | 12.56 | 21.68 | 23.13 |
| Gembloux | Culicoides | 07 Aug 2012 | 8.19 | 30.9 | 30.81 |
| Avaritia | 07 Aug 2012 | 9.54 | 31.1 | 31.51 | |
| Libramont | Culicoides | 08 Aug 2012 | 15.05 | 33.96 | 40.0* |
| Culicoides | 08 Aug 2012 | 14.98 | 34.2 | 38.8* | |
| Avaritia | 08 Aug 2012 | 17.62 | 34.28 | 40.0* | |
| Avaritia | 08 Aug 2012 | 17.27 | 22.12 | 23.6 | |
| Avaritia | 08 Aug 2012 | 17.14 | 33.46 | 37.0* | |
| Avaritia | 08 Aug 2012 | 17.22 | 35.06 | 40.0* | |
| Avaritia | 08 Aug 2012 | 17.92 | 35.01 | 40.0* | |
| Avaritia | 08 Aug 2012 | 16.73 | 35.84 | 40.0* | |
| Avaritia | 08 Aug 2012 | 17.91 | 35.06 | 40.0* | |
| Culicoides | 23 Aug 2012 | 9.56 | 33.54 | 38.76* | |
| Avaritia | 23 Aug 2012 | 10.51 | 21.71 | 23.96 | |
| Avaritia | 23 Aug 2012 | 10.7 | 21.02 | 23.04 | |
| Avaritia | 23 Aug 2012 | 10.22 | 31.94 | 38.03* | |
| Avaritia | 23 Aug 2012 | 10.3 | 33.33 | 37.98* | |
| Avaritia | 23 Aug 2012 | 10.88 | 21.05 | 23.64 | |
| Avaritia | 23 Aug 2012 | 11.8 | 30.34 | 36.35* | |
| Avaritia | 23 Aug 2012 | 10.56 | 33.24 | 40.0* | |
| Avaritia | 23 Aug 2012 | 10.93 | 20.99 | 22.26 | |
| Avaritia | 23 Aug 2012 | 11.31 | 21.54 | 23.89 | |
| Avaritia | 23 Aug 2012 | 10.86 | 19.35 | 21.0 | |
| Avaritia | 23 Aug 2012 | 10.36 | 33.07 | 38.79* | |
| Avaritia | 23 Aug 2012 | 10.32 | 33.22 | 40.0* | |
| Avaritia | 23 Aug 2012 | 10.92 | 19.91 | 21.53 | |
tested in two-step rRT-PCR.
I.C. = internal control.
However, the infection prevalences in August in the other regions were however clearly lower (0.4, 0.3, and 0.2% in Avaritia in Antwerp, Liège and Gembloux, respectively, and 0.4% in Culicoides in Gembloux). In those regions, non-protected sheep and calves borne after the 2011 epidemic have probably allowed the replication of the virus and served as a source for SBV infection of the midges.
The sudden appearance of SBV infected midges in all studied regions in August raises the question as to where the virus came from. A first possibility could be that the virus had overwintered in either its host or its vector. Since only a short viraemia occurs in sheep and cattle [1], [24], [25], [36], [37], it seems unlikely that the virus overwintered in these animals. Overwintering in another, so far unidentified, animal host could be another possibility. When overwintering in Culicoides is considered, this could either occur in the low number of adult midges that are capable to survive Belgian winter conditions [38] or in infected eggs or larval stages after transovarial virus transmission. To get a first idea if overwintering in the vector had occurred by transovarial virus transmission in Belgium, 69 pools representing 1,359 nulliparous females caught in May 2012 at places where SBV had circulated in 2011 were tested (Table 3). The fact that all were found negative provides an indication that transovarial transmission is not likely to occur. This should, however, be further investigated since it was recently reported that SBV RNA was detected in midges considered as nulliparous based on visual inspection in Poland in 2012 [18]. Laboratory based studies will probably be necessary to unambiguously show whether transovarial transmission in biting midges occurs or not. Another possibility that cannot be excluded at the moment is that, besides Culicoides, other hematophagous insects can function as (overwintering) vectors for SBV. In this respect, no SBV has been found in 868 hibernating mosquitoes in the Netherlands collected from January to March 2012 [39]. Although no SBV positive insect vectors have been detected during winter yet, the finding that sheep became viremic for SBV in the beginning of January 2013 in Germany during a period characterized by a rise of the minimum temperature above 5°C supports the hypothesis that SBV overwinters in hematophagous insects [40]. Alternatively to overwintering in hosts or vectors, the observed presence of SBV in 2012 in Belgium could also have been the result of a reintroduction of the virus by arthropods coming in from neighbouring countries since SBV circulation in 2012 has been reported in France, Germany and The Netherlands [16], [26], [29].
Table 3. Schmallenberg virus detection in nulliparous female midges caught in May 2012.
| Antwerp | Gembloux | # SBV pos | |
| Avaritia | 24 (476) | 31 (620) | 0 |
| Culicoides | 5 (85) | – | 0 |
| Monoculicoides | 8 (160) | – | 0 |
| Other species | 1 (18) | – | 0 |
| Total | 38 (739) | 31 (620) | 0 |
Similar to the sudden appearance of SBV in Culicoides in August 2012, the abrupt absence of SBV in examined pools of Culicoides caught in September 2012 and later is remarkable (Figure 2). Based on the knowledge of the fast spread of SBV during the 2011 epidemic, this is most probably caused by the rapid infection of all residual non-protected animals, associated with the induction of a protective immune response that prevented further spread.
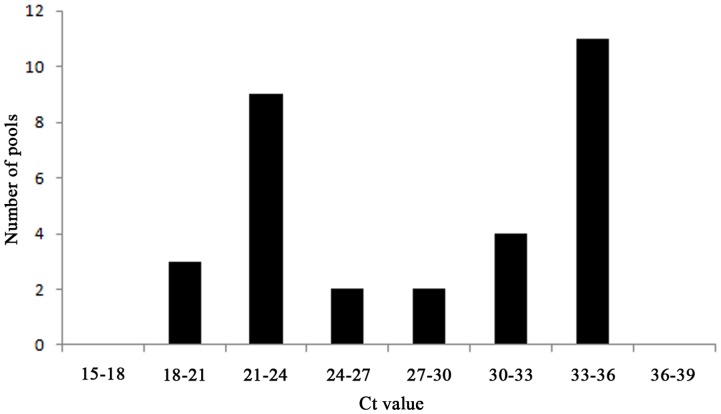
Due to the outbreak of bluetongue virus in 2006 in Central and Northern Europe [41] and the recent SBV emergence, scientists have been confronted with two viruses that were immediately suspected to be spread by biting midges once first diagnosis in animal hosts had occurred. In the urge to identify responsible vector species as fast as possible, several Culicoides species have been proposed as putative vectors based on the identification of the virus in pools of field caught Culicoides by rRT-PCR [14], [17]–[19], [35], [42]–[44]. Several aspects related to (1) the diagnostic technique, (2) arbovirus replication and dissemination characteristics in their vectors and (3) pool composition, however, cause that such results should be interpreted with caution, particularly if only a limited number of positive pools are found [20], [45]. Two main obstacles are that i) rRT-PCR also detects non-infectious viral RNA, making it uncertain that obtained Ct values truly represent virus capable of infecting new hosts, and ii) rRT-PCR positive whole midges do not necessarily represent vectors producing transmissible virus since barriers preventing virus to enter or escape from the midgut and preventing virus dissemination through the haemocoel exist in Culicoides that may prevent dissemination of the virus to the head and salivary glands after uptake of an infectious blood meal [46], [47]. Upon the emergence of SBV, these issues were partially addressed by some studies that attempted detecting SBV in species-specific pools consisting only of Culicoides’ heads [13], [15] since this implicates that rRT-PCR positive pools contain most probably fully disseminated infections. However, this method is labor intensive and therefore offers only chance to detect virus positive Culicoides in the restricted number of pools tested when vector competence for the studied virus is high [20]. Recently, a new approach to evaluate arbovirus dissemination via rRT-PCR in field caught whole Culicoides was proposed based on the results obtained with laboratory grown C. sonorensis midges orally fed with SBV or BTV-infected blood via a membrane system [20], [30]. It was found that SBV infection of a susceptible vector species resulted in a bimodal distribution of the obtained Ct values. Ct values close to the first peak indicate a fully disseminated infection and presence of transmissible virus while Ct values close to the second peak indicate sub-transmissible infections. SBV infection of refractory species (C. nubeculosus) did not result in a bimodal distribution of obtained Ct values. When a similar data analysis was applied to the Ct values obtained in this study for the 31 SBV-S segment positive pools of Avaritia (Table 2), a bimodal distribution could be found with a first peak of Ct values between 21 and 24 and a second peak with Ct values between 33 and 36 (Figure 3). Following the interpretation of the proposed approach [20], this result predicts that the subgenus of Avaritia contains competent SBV vector species. This is in line with previous publications suggesting Obsoletus complex, C. chiopterus and C. dewulfi midges as putative vectors based on rRT-PCR analysis of species-specific pools of heads [13], [15] and therefore represents a field validation of the proposed approach.
Figure 3. Ct value frequency distribution of Schmallenberg virus positive pools of Avaritia.
The necessity for a relatively high number of rRT-PCR positive midges of a certain species to reliably estimate if a bimodal distribution is present will probably sometimes limit the applicability of this approach to assess the vectorial competence of field caught midges. In the present study, 4 SBV-S positive pools containing midges of the subgenus Culicoides were found with Ct values for the SBV-S segment between 30.9 and 34.2 (Table 2). This number is clearly not sufficient to assess if species belonging to the subgenus Culicoides are refractory species to SBV or if they (or a specific species of the subgenus Culicoides) are susceptible and the pool contained individuals with sub-transmissible amounts of SBV. Similar inconclusive results were obtained by rRT-PCR analysis of pools of heads of C. pulicaris since these were positive for the S segment of SBV but not the L segment [13]. It seems advisable to interpret these data in a conservative way and not to propose C. pulicaris as a competent vector at this moment.
Acknowledgments
We thank Marie-Rose Bollen and Hughes Seutin for their help with the collection and identification of the midges and the preparation of the pools. We thank Virginie Colasse for the excellent help with rRT-PCR testing of Culicoides and Flavien Riocreux for the preparation of the map with Culicoides trapping locations.
Funding Statement
Monitoring of Culicoides in Belgium in 2012 was financed by the Belgian Federal Agency for the Safety of the Food Chain (FAVV-AFSCA). rRT-PCR analysis of Culicoides for presence of SBV was funded by the Federal Public Service of Health, Food Chain Safety and Environment (RF12/6270) and the European Union as outlined in Council Decision 2012/349/EU concerning a financial contribution by the Union for studies on Schmallenberg virus. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, et al. (2012) Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis 18: 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garigliany MM, Bayrou C, Kleijnen D, Cassart D, Jolly S, et al. (2012) Schmallenberg virus: a new Shamonda/Sathuperi-like virus on the rise in Europe. Antiviral Res 95: 82–87. [DOI] [PubMed] [Google Scholar]
- 3. Herder V, Wohlsein P, Peters M, Hansmann F, Baumgärtner W (2012) Salient lesions in domestic ruminants infected with the emerging so-called Schmallenberg virus in Germany. Vet Pathol 49: 588–591. [DOI] [PubMed] [Google Scholar]
- 4. Van den Brom R, Luttikholt SJ, Lievaart-Peterson K, Peperkamp NH, Mars MH, et al. (2012) Epizootic of ovine congenital malformations associated with Schmallenberg virus infection. Tijdschr Diergeneeskd 137: 106–111. [PubMed] [Google Scholar]
- 5.Chaintoutis SC, Kiossis E, Giadinis ND, Brozos CN, Sailleau C, et al.. (2013) Evidence of Schmallenberg virus circulation in ruminants in Greece. Trop Anim Health Prod In press. [DOI] [PubMed]
- 6.Conraths FJ, Peters M, Beer M (2013) Schmallenberg virus, a novel orthobunyavirus infection in ruminants in Europe: potential global impact and preventive measures. N Z Vet J 61, 63–67. [DOI] [PubMed]
- 7. Doceul V, Lara E, Sailleau C, Belbis G, Richardson J, et al. (2013) Epidemiology, molecular virology and diagnostics of Schmallenberg virus, an emerging orthobunyavirus in Europe. Vet Res 44: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Food Safety Authority: “Schmallenberg” virus: analysis of the epidemiological data (2013) Supporting Publications 2013: EN-429[22pp.]. Available: http://www.efsa.europa.eu/en/efsajournal/doc/429e.pdf Accessed 2013 Dec 24.
- 9. Al-Busaidy SM, Mellor PS (1991) Isolation and identification of arboviruses from the Sultanate of Oman. Epidemiol Infect 106: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bryant JE, Crabtree MB, Nam VS, Yen NT, Duc HM, et al. (2005) Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am J Trop Med Hyg 73: 470–473. [PubMed] [Google Scholar]
- 11. St George TG, Standfast HA, Cybinski DH (1978) Isolations of akabane virus from sentinel cattle and Culicoides brevitarsis . Aust Vet J 54: 558–561. [DOI] [PubMed] [Google Scholar]
- 12. Yanase T, Kato T, Kubo T, Yoshida K, Ohashi S, et al. (2005) Isolation of bovine arboviruses from Culicoides biting midges (Diptera : Ceratopogonidae) in southern Japan: 1985–2002. J Med Entomol 42: 63–67. [DOI] [PubMed] [Google Scholar]
- 13. De Regge N, Deblauwe I, De Deken R, Vantieghem P, Madder M, et al. (2012) Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transbound Emerg Dis 59: 471–475. [DOI] [PubMed] [Google Scholar]
- 14. Rasmussen LD, Kristensen B, Kirkeby C, Rasmussen TB, Belsham GJ, et al. (2012) Culicoides as vectors of Schmallenberg virus. Emerg Infect Dis 18: 1204–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elbers AR, Meiswinkel R, van Weezep E, van Oldruitenborgh-Oosterbaan MM, Kooi EA (2013) Schmallenberg virus in Culicoides spp. biting midges, the Netherlands, 2011. Emerg Infect Dis 19: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbers AR, Meiswinkel R, van Weezep E, Kooi EA, van der Poel WH (2013) Schmallenberg Virus in Culicoides Biting Midges in the Netherlands in 2012. Transbound Emerg Dis, In press, doi: 10.1111/tbed.12128. [DOI] [PubMed]
- 17. Goffredo M, Monaco F, Capelli G, Quaglia M, Federici V, et al. (2013) Schmallenberg virus in Italy: a retrospective survey in Culicoides stored during the bluetongue Italian surveillance program. Prev Vet Med 111: 230–236. [DOI] [PubMed] [Google Scholar]
- 18. Larska M, Lechowski L, Grochowska M, Zmudziński JF (2013) Detection of the Schmallenberg virus in nulliparous Culicoides obsoletus/scoticus complex and C. punctatus-The possibility of transovarial virus transmission in the midge population and of a new vector. Vet Microbiol 166: 467–473. [DOI] [PubMed] [Google Scholar]
- 19. Larska M, Polak MP, Grochowska M, Lechowski L, Związek JS, et al. (2013) First report of Schmallenberg virus infection in cattle and midges in Poland. Transbound Emerg Dis 60: 97–101. [DOI] [PubMed] [Google Scholar]
- 20. Veronesi E, Henstock M, Gubbins S, Batten C, Manley R, et al. (2013) Implicating Culicoides biting midges as vectors of Schmallenberg virus using semi-quantitative RT-PCR. PLoS One 8(3): e57747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Méroc E, De Regge N, Riocreux F, Caij AB, van den Berg T, et al.. (2013) Distribution of Schmallenberg Virus and Seroprevalence in Belgian Sheep and Goats. Transbound Emerg Dis, In press ahead, doi: 10.1111/tbed.12050. [DOI] [PubMed]
- 22. Méroc E, Poskin A, Van Loo H, Quinet C, Van Driessche E, et al. (2013) Large-scale cross-sectional serological survey of schmallenberg virus in belgian cattle at the end of the first vector season. Transbound Emerg Dis 60: 4–8. [DOI] [PubMed] [Google Scholar]
- 23. Linden A, Desmecht D, Volpe R, Wirtgen M, Gregoire F, et al. (2012) Epizootic spread of Schmallenberg virus among wild cervids, Belgium, Fall 2011. Emerg Infect Dis 18: 2006–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wernike K, Eschbaumer M, Schirrmeier H, Blohm U, Breithaupt A, et al. (2013) Oral exposure, reinfection and cellular immunity to Schmallenberg virus in cattle. Vet Microbiol 165: 155–159. [DOI] [PubMed] [Google Scholar]
- 25. Wernike K, Nikolin VM, Hechinger S, Hoffmann B, Beer M (2013) Inactivated Schmallenberg virus prototype vaccines. Vaccine 31: 3558–3563. [DOI] [PubMed] [Google Scholar]
- 26. Sailleau C, Bréard E, Viarouge C, Desprat A, Doceul V, et al. (2012) Acute Schmallenberg virus infections, France. Emerg Infect Dis 19: 321–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bayrou C, Garigliany MM, Cassart D, Jolly S, Desmecht D (2013) Schmallenberg virus circulation in Belgium in 2012. Vet Rec 172: 296. [DOI] [PubMed] [Google Scholar]
- 28. Claine F, Coupeau D, Wiggers L, Muylkens B, Kirschvink N (2013) Schmallenberg Virus among Female Lambs, Belgium, 2012. Emerg Infect Dis 19: 1115–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Conraths FJ, Kämer D, Teske K, Hoffmann B, Mettenleiter TC, et al. (2013) Reemerging Schmallenberg virus infections, Germany, 2012. Emerg Infect Dis 19: 513–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Veronesi E, Antony F, Gubbins S, Golding N, Blackwell A, et al. (2013) Measurement of the infection and dissemination of bluetongue virus in Culicoides biting midges using a semi-quantitative rt-PCR assay and isolation of infectious virus. PLoS One 8(8): e70800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venter GJ, Labuschagne K, Hermanides KG, Boikanyo SNB, Majatladi DM, et al. (2009) Comparison of the efficiency of five suction light traps under field conditions in South Africa for the collection of Culicoides species. Vet Parasitol 166: 299–307. [DOI] [PubMed] [Google Scholar]
- 32.Delécolle JC (1985) Nouvelle contribution à l’étude systématique et iconographique des espèces du genre Culicoides (Diptera : Ceratopogonidae) du nord-est de la France. PhD Thesis, University of Strasbourg, Strasbourg.
- 33. Fassotte C, Delécolle JC, Cors R, Defrance T, De Deken R, et al. (2008) Culicoides trapping with Rothamsted suction traps before and during the bluetongue epidemic of 2006 in Belgium. Prev Vet Med 87: 74–83. [DOI] [PubMed] [Google Scholar]
- 34. Bilk S, Schulze C, Fischer M, Beer M, Hlinak A, et al. (2012) Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet Microbiol 159: 236–238. [DOI] [PubMed] [Google Scholar]
- 35. Vanbinst T, Vandenbussche F, Vandemeulebroucke E, De Leeuw I, Deblauwe I, et al. (2009) Bluetongue virus detection by real-time RT-PCR in Culcoides captured during the 2006 epizootic in Belgium and development of an internal control. Transbound Emerging Dis 56: 170–177. [DOI] [PubMed] [Google Scholar]
- 36. Wernike K, Eschbaumer M, Breithaupt A, Hoffmann B, Beer M (2012) Schmallenberg virus challenge models in cattle: infectious serum or culture-grown virus? Vet Res 43: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wernike K, Hoffmann B, Bréard E, Bøtner A, Ponsart C, et al. (2013) Schmallenberg virus experimental infection of sheep.Vet Microbiol. 166: 461–466. [DOI] [PubMed] [Google Scholar]
- 38. Losson B, Mignon B, Paternostre J, Madder M, De Deken R, et al. (2007) Biting midges overwintering in Belgium. Vet Rec 160: 451–452. [DOI] [PubMed] [Google Scholar]
- 39.Scholte EJ, Mars MH, Braks M, Den Hartog W, Ibañez-Justicia A, et al.. (2013) No evidence for the persistence of Schmallenberg virus in overwintering mosquitoes. Med Vet Entomol, In press, doi: 10.1111/mve.12010. [DOI] [PubMed]
- 40. Wernike K, Kohn M, Conraths F, Werner D, Kameke D, et al. (2013) Transmission of Schmallenberg virus during winter, Germany. Emerg Infect Dis 19: 1701–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson AJ, Mellor PS (2009) Bluetongue in Europe: past, present and future. Philos Trans R Soc Lond B Biol Sci 364: 2669–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mehlhorn H, Walldorf V, Klimpel S, Jahn B, Jaeger F, et al. (2007) First occurence of Culicoides obsoletus-transmitted Bluetongue virus epidemic in Central Europe. Parasitol Res 101: 219–228. [DOI] [PubMed] [Google Scholar]
- 43. Meiswinkel R, Van Rijn P, Leijs P, Goffredo M (2007) Potential new Culicoides vector of bluetongue virus in northern Europe. Vet Rec 161: 564–565. [DOI] [PubMed] [Google Scholar]
- 44. Dijkstra E, van der Ven IJ, Meiswinkel R, Hölzel DR, Van Rijn PA, et al. (2008) Culicoides chiopterus as a potential vector of bluetongue virus in Europe. Vet Rec 162: 422. [DOI] [PubMed] [Google Scholar]
- 45. Carpenter S, McArthur C, Selby R, Ward R, Nolan DV, et al. (2008) Experimental infection studies of UK Culicoides species midges with bluetongue virus serotypes 8 and 9. Vet Rec 163: 589–592. [DOI] [PubMed] [Google Scholar]
- 46. Mellor PS (2000) Replication of arboviruses in insect vectors. J Comp Pathol 123: 231–247. [DOI] [PubMed] [Google Scholar]
- 47. Mellor PS, Boorman J, Baylis M (2000) Culicoides biting midges: Their role as arbovirus vectors. Annu Rev Entomol 45: 307–340. [DOI] [PubMed] [Google Scholar]