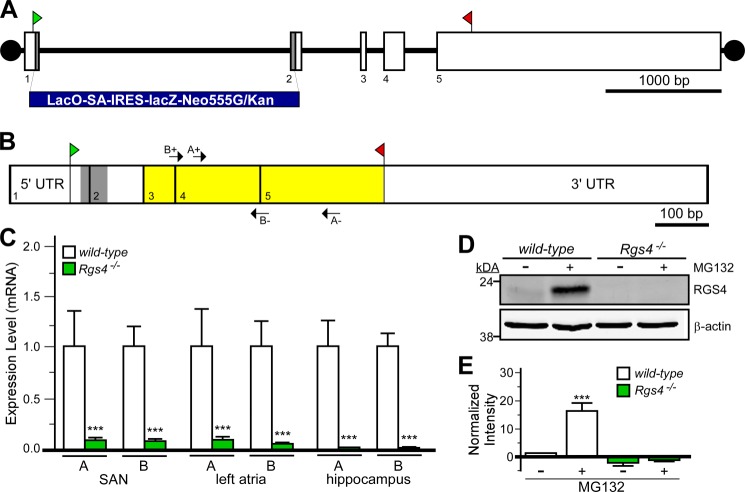
FIGURE 1.
Characterization of Rgs4−/− mice. A, schematic depiction of the Rgs4 gene, which consists of five exons (boxes 1–5). The translation initiation (ATG) and termination (STOP) codons are shown in green and red, respectively. The gray regions in exons 1 and 2 (along with intervening intronic sequence) were replaced with the lacZ-containing cassette (LacO-SA-IRES-lacZ-Neo555G/Kan) to generate B6;129P2-Rgs4tm1Dgen/J mice. B, schematic depiction of RGS4 mRNA, with exon boundaries denoted by vertical lines and numbers. The gray domain corresponds to the 58-bp fragment of coding sequence missing in the RGS4 mutant mice. Yellow highlighting shows the location of coding sequence for the catalytic (RGS) domain of RGS4. Arrows denote the positions and identities of the primer sets (A+/− and B+/−) used for quantitative RT-PCR. C, quantitative RT-PCR analysis of RGS4 expression in cardiac tissues (SAN and left atria) and hippocampi from wild-type and B6;129P2-Rgs4tm1Dgen/J mice using the primer sets depicted in C. Expression levels were normalized within samples to GAPDH (the levels of which were comparable in all tissues examined) and to wild-type samples for each primer set. ***, p < 0.001 versus wild-type mice (within tissue and primer set by t test). D, immunoblotting for RGS4 in primary hippocampal cultures (10 days in vitro) from wild-type and B6;129P2-Rgs4tm1Dgen/J mice. Cultures were pretreated with MG132 (50 μm) for 6 h prior to protein isolation. E, quantification of RGS4 immunoblotting data (n = three separate experiments). A significant impact of group was observed (F3,11 = 29.8; p < 0.001). ***, p < 0.001 versus wild-type mice (untreated).