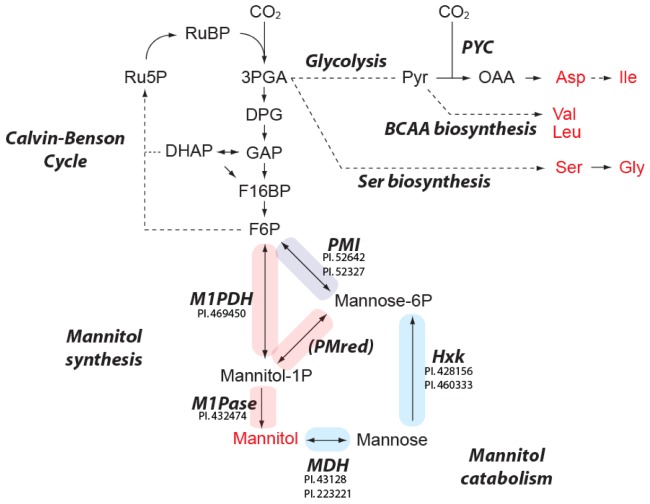
Figure 3.
Overview of the CO2 fixation pathway and predicted enzymes for mannitol metabolism in E. huxleyi. Regular and bold italic indicate metabolites and enzymes or metabolic pathways, respectively. Metabolites in red were detected in the current study. Other metabolites except for pyruvate and mannose are hardly detected by GC-MS analysis. The proteins showing significant homology to known enzymes are listed close to the enzyme name. Protein IDs are taken from JGI Emiliania huxleyi CCMP1516 main genome assembly v1.0. Solid and dashed arrows show single and multiple reactions, respectively. The reactions colored by red, blue and purple are related to manntiol biosynthesis, catabolism and both pathways, respectively. Ru5P, riburose-5-phosphate; RuBP, riburose bisphosphate; 3PGA, 3-phosphoglycerate; DPG, 1,3-diphosphoglycerate; GAP, glyceraldehyd-3-phosphate; DHAP, dihydroxyacetone phosphate; F16BP, fructose-1,6-bisphosphate; F6P, fructose-6-phosphate; Pyr, pyruvate; OAA, oxaloacetate; BCAA, branched chain amino acid; PYC, pyruvate carboxylase; PMI, phosphomannose isomerase; PMred, phosphomannose reductase; M1PDH, mannitol-1-phosphate dehydrogenase; M1Pase, mannose-1-phosphatate; MDH, mannitol dehydrogenase; Hxk, hexokinase.