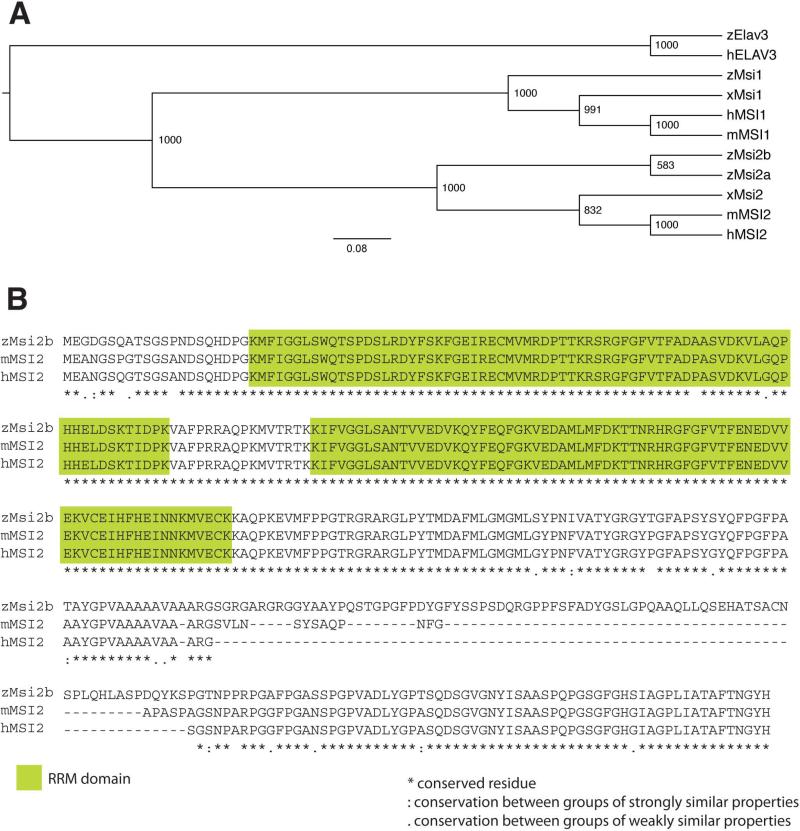
Figure 2. Comparison among members of the Msi family of RNA-binding proteins.
(A) Phylogenetic tree of Musashi proteins in vertebrates was generated by ClustalX analysis. Human, hMSI1 and hMSI2; mouse, mMSI1 and mMSI2; Xenopus, xMsi1 and xMsi2 and Zebrafish, zMsi1, zMsi2a, and zMsi2b. Zebrafish and human RNA-binding protein, zElav3 and hELAV3, were included in the analysis as outgroups. In zebrafish, Msi2 has diverged further into zMsi2a and zMsi2b. The numbers on the nodes of the tree represent the bootstrap values from Neighbor joining analysis.
(B) Alignment of zMsi2b protein sequence with mouse and human MSI2. The RNA-binding domains (green boxes) are highly conserved between zMsi2b and mammalian MSI2, with only two amino acids substitutions in the first domain. Sequence divergences consist of an additional sequence of amino acids between positions 263 and 343 of zMsi2b sequence. Full conservation of residues are indicated by asterisk (*); “:” and “.” indicate full conservation of ‘strong’ and ‘weaker’ residues, respectively.