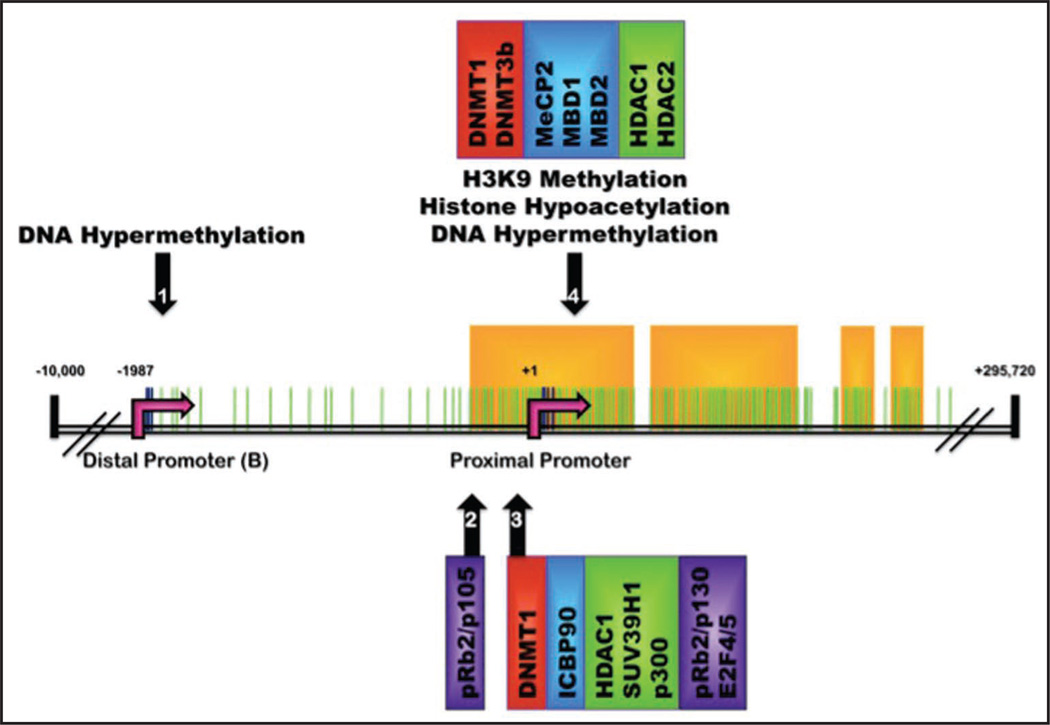
Figure 3.
Methylation sites and epigenetic complexes identified at the estrogen receptor α promoter: Schematic of the ERα gene (ESR1) on chromosome 6q25.1. CpG dinucleotides located on ERα promoter region from −2000 to +2000 are depicted by green lines. Specific CpG residues demonstrated to be sites of potential CpG methylation are marked by dark blue lines and are situated at the distal (B) and proximal promoters (identified by pink arrows at −1987 and +1 respectively). Yellow boxes designate predicted CpG islands (as defined by www.urogene.org/methprimer) which identify four regions of dense CpG population located at the proximal promoter (nucleotides −280 to +359) and just upstream of the proximal promoter (residing at nucleotides +443 to +1140; +1380 to +1427; and +1463 to +1563). Promoter regions determined to contain specific DNA and histone modifications are marked by numbered black arrows. Epigenetic regulators associated with each specific promoter region include DNA Methyltransferases (red boxes), Methyl-CpG-Binding Proteins (blue boxes), histone modifying enzymes (green boxes) and other transcriptional repressors (purple boxes).