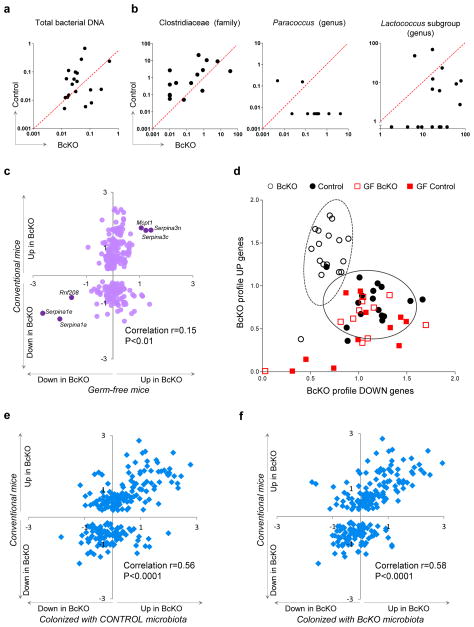
Figure 3.
Microbiota are necessary for intestinal alterations in BcKO mice. (a) Amount of total bacterial DNA (ng) per 10 ng of isolated total DNA from jejunum content of BcKO and control mice (each dot represents data from two mice of the same litter. (b) Amount of DNA (pg) for Clostridiacea (family), Paraccococus (genus) and for an operational taxonomic unit corresponding to a subgroup of Lactococcus (genus) per 10 ng of isolated total DNA from jejunum content of BcKO and control mice. Data represented as in a. (c) Gene expression ratios between BcKO and control (WT and heterozygous) (y axis; n = 27 per group) and between germ-free BcKO versus germ-free control (WT and heterozygous) mice (x axis; n = 11 per group). Each symbol represents one gene. Six genes that are differentially expressed between germ-free BcKO and germ-free control mice are labeled. (d) Two-dimensional visualization of the gene expression of upregulated (BcKO profile up) and downregulated (BcKO profile down) genes in four groups of mice. Each symbol represents one mouse. Values are summary metrics of upregulated (y axis) and downregulated (x axis) genes of the BcKO profile. Ellipses were represent two s.d. from the centroids for BcKO (dashed) and Control (black) groups ]. (e) Gene expression ratios between conventional BcKO and control (WT and heterozygous) mice (y axis; n = 27 per group) and between ex–germ-free BcKO versus control mice colonized with microbiota from control mice (x axis; n = 8 per group). (f) Gene expression ratios between conventional BcKO and control (WT and heterozygous) mice (y axis; n = 27 per group) and between ex–germ-free BcKO versus control mice colonized with microbiota from control mice (x axis; n = 5 per group). Each symbol represents one gene in e and f. Unless otherwise indicated, the control mice were heterozygotes.