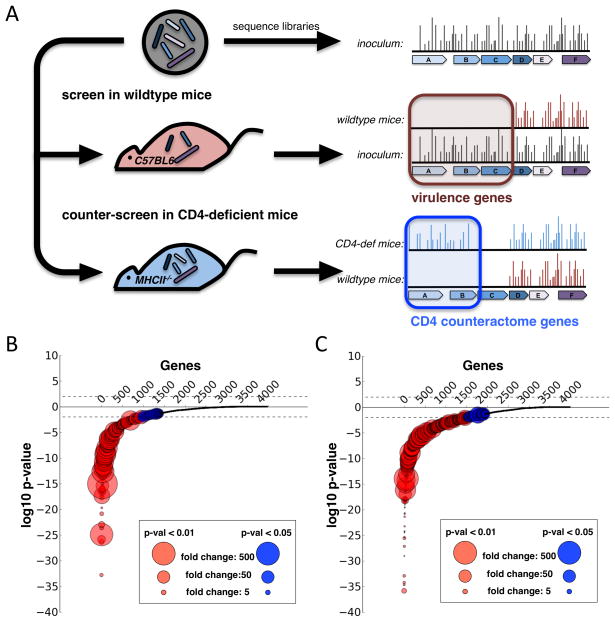
Figure 1.
A. MHC Class II KO and wild type mice were infected with 106 bacilli from our transposon library. At 10 days and 45 days after infection, 4 mice in each group were sacrificed and spleen homogenates were plated to recover surviving bacteria. To discover genes required for surviving CD4 T Cell immunity, we searched for genes that were required for growth in a wild type mouse but not required in the MHC Class II mice. For both the d10 (B.) and d45 (C.) timepoints, we calculated for each gene the insertion count differences between the input library and the recovered libraries from wild type mice, and a p-value expressing the significance of this differences. We then log-transformed the p-values and gave each gene with a loss of insertions a negative log-p-value and each gene with a gain of insertions a positive log-p-value. After ordering the genes based on their p-values, we plotted each gene as a dot with log-p-values on the y-axis and the size of the circles representing the fold-change of insertion count differences.