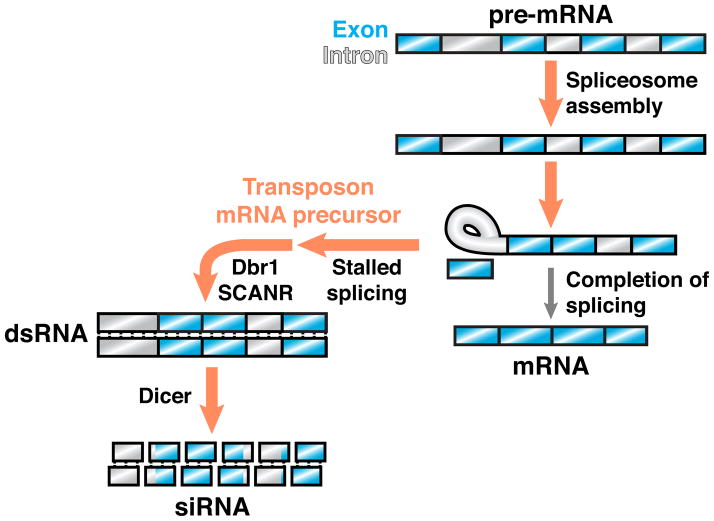
Figure 2. Stalled spliceosomes license RNA silencing in an endo-siRNA pathway.
In the yeast C. neoformans, transcripts targeted by RNA silencing, which primarily include transposons, exhibit sequence features predictive of poor splicing and tend to stall in spliceosomes. The stalled splicing of transposon mRNA precursors is required for siRNA biogenesis mediated by SCANR, a protein complex that contains an RdRP and physically associates with the spliceosome. Lariat debranching enzyme (Dbr1) is also required for siRNA production, suggesting that transposon mRNA precursors in the lariat intermediate stage are linearized to enable dsRNA formation by SCANR. In this hypothetical example, splicing of a transcript’s first intron stalls at the lariat intermediate stage, whereas downstream introns remain partially spliced.