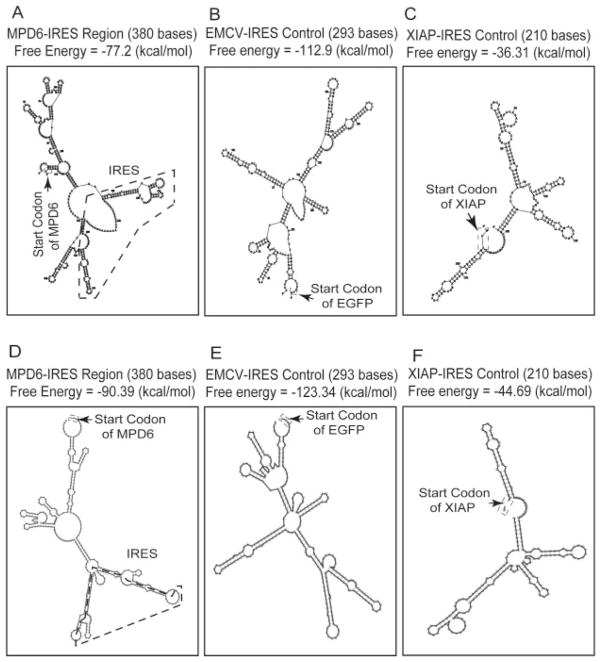
FIGURE 2.
The predicted stem-loop structure in MPD6-IRES. The cis-acting regulatory elements in 3′-UTR were analyzed using the IRES website (〈http://ifr31w3.toulouse.inserm.fr/IRESdatabase/〉) and the UTR website (〈www2.ba.itb.cnr.it/UTRSite/〉) with generous support by Dr. S. Liuni at the Bioinformatics and Genomic Group in Italy. The secondary structures of MPD6-IRES (A and D), EMCV-IRES (B and E), and XIAP-IRES (C and F) were predicted by using two web-based algorithms, MFOLD-Zuker (〈www.bioinfo.rpi.edu/applications/mfold/old/rna/〉) (A–C) and RNAfold (〈http://rna.tbi.univie.ac.at/〉) (D–F). The free energy of the secondary structure of these IRES regions was also calculated with both algorithms. The start codon and IRES region of MPD6 are indicated.