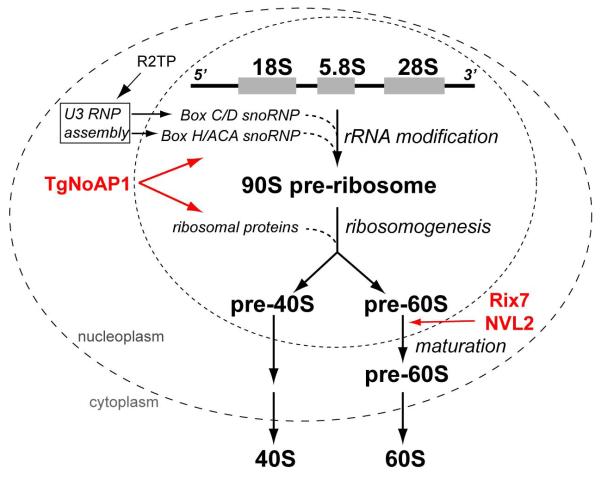
Figure 9. Subcellular localization of the complexes involved in eukaryotic ribosome biogenesis.
The multi-stage process of ribosome component synthesis and assembly takes place in three cellular compartments: nucleolus, nucleus and cytoplasm. While rDNA is transcribed in the fibrillar core of the nucleolus, the critical complexes (U3 snoRNP) assisting co-transciptional modifications of pre-rRNA (box C/D RNA complex catalyzes 2′-O-methylation and H/ACA RNA complex promotes pseudouridine modification) are assembled and translocated from nucleoplasm to nucleolus under control of the R2TP complexes. Ribosomogenesis continues in the nucleolus where maturation of the 90S pre-ribosome particle leads to release of pre-40S and pre-60S ribosomal subunits into the nucleus. The 60S large subunit undergoes additional processing in the nucleoplasm; a step facilitated by the yeast (Rix7) and human (NVL2) orthologs of TgNoAP1. Mature individual small (40S) and large (60S) subunits of the ribosome are released into the cytoplasm. Evidence presented in this study strongly indicates that Toxoplasma TgNoAP1 associates with the complexes involved in modification of the pre-rRNA and maturation of the 90S preribosome, which is upstream in ribosome biogenesis from the 60S maturation steps that acquire closely related AAA ATPases, Rix7 and NVL2.