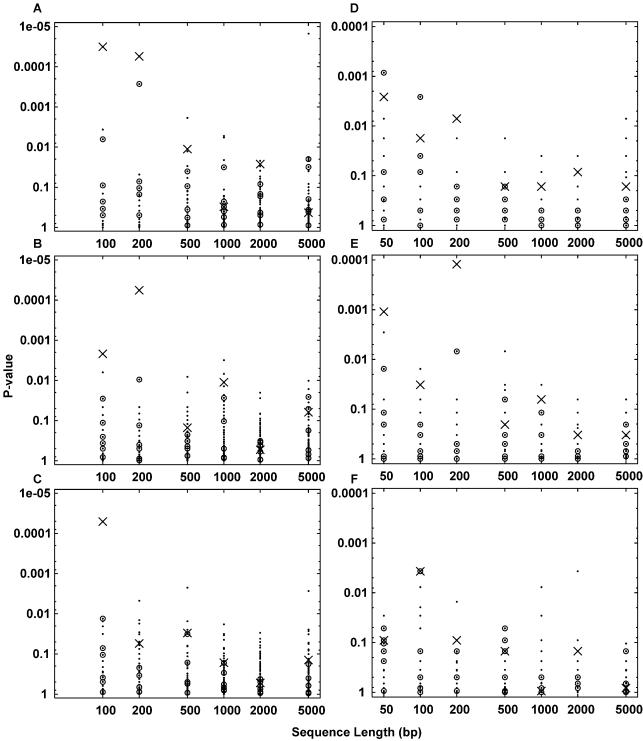
Figure 4.
Detection by contingency table based methods of EREs embedded in random DNA sequences of varying length. In all panels, the P-values of the 108 Jaspar motifs are plotted as dots. Crosses indicate the PPARγ motif, and circles indicate the six other ERE-like nuclear receptor motifs. (A, B, C) Motif counting method. Length 50 sequences were not analyzed because the number of possible locations is <1000 for some motifs, making the 0.1% threshold criterion impossible. (D, E, F) Sequence counting method. (A, D) Results for 15 ERE-containing sequences with no decoy sequences. (B, E) Results for 15 ERE-containing sequences with five decoy sequences. (C, F) Results for 15 ERE-containing sequences with 15 decoy sequences.