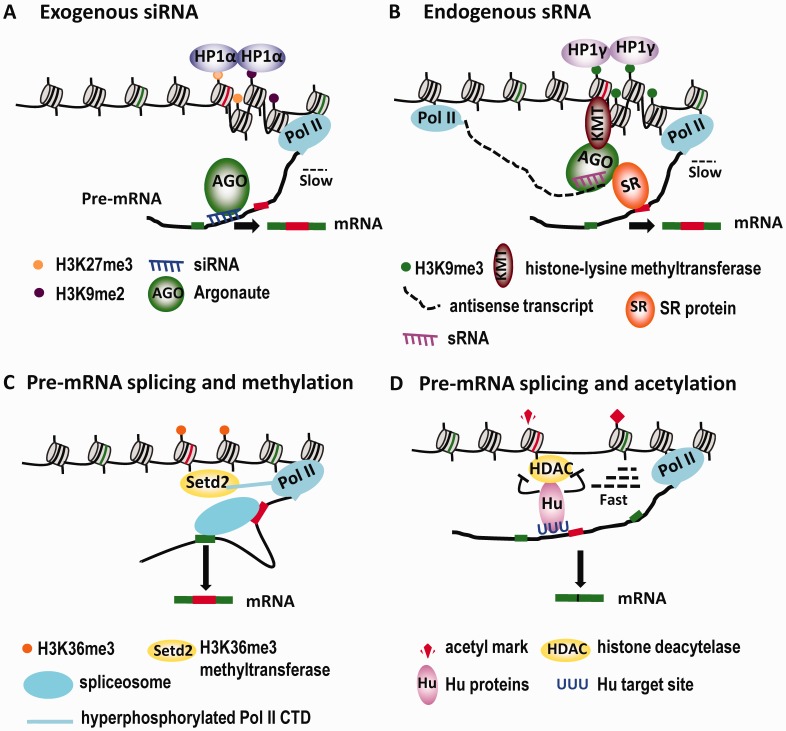
Figure 2.
RNA-guided mechanisms that link chromatin modification and alternative splicing. (A) Treatment of cells with siRNAs complementary to regions surrounding an alternative exon induces deposition of the heterochromatin marks H3K9me2/H3K27me3 and H3K9me3 at the target site, which are bound by HP1α and HP1γ, respectively. This results in a reduction of the Pol II elongation rate and increased exon inclusion. (B) An intragenic antisense transcript generates sRNAs that target the intron–exon junction region close to the variable exons of CD44, which leads to enrichment of the H3K9me3 mark and recruitment of HP1γ. Consequently, the Pol II elongation rate is reduced and variable exons are included at a higher level. In addition, the AGO protein that is associated with the sRNA likely recruits multiple splicing factors through direct interactions, which further contributes to the increased inclusion of variable exons (see text for details). (C) The process of pre-mRNA splicing reinforces the coincident distribution of Setd2 (the mammalian ortholog of yeast Set2), the H3K36me3 methyltransferase and H3K36me3 marks on exons in genes undergoing active transcription. (D) Hu proteins bound at their target pre-mRNA sites interact with and inhibit the enzymatic activity of HDAC2, leading to localized H3 and H4 hyperacetylation in the corresponding chromosomal region. The resulting increase in the Pol II elongation rate through this region promotes skipping of the Hu-regulated alternative exons.