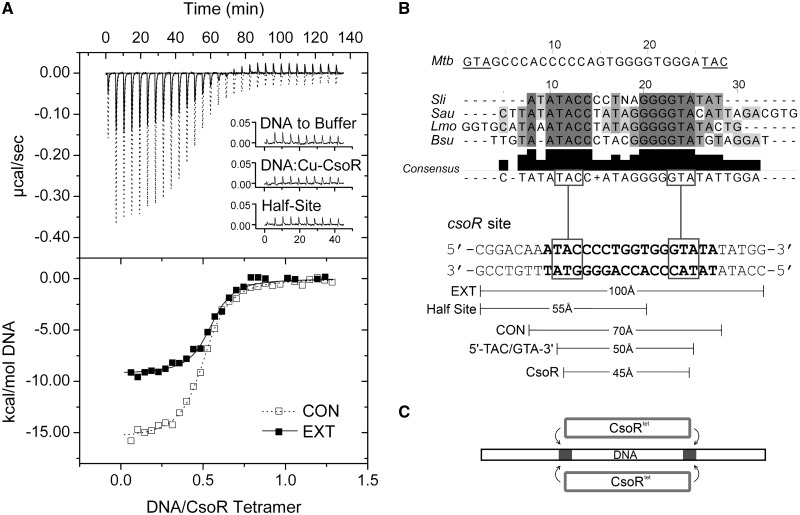
Figure 2.
Thermodynamics and operator site symmetry. (A) ITC binding profiles at 25°C and fits to a one set of sites binding model (dashed and solid line) for wild-type CsoRSl with the consensus (CON) and extended (EXT) csoR operator (Table 1). A null response similar to DNA dilutions (inset) is observed with Cu(I)-CsoR, as well as with a construct comprising half the csoR-EXT sequence (Table 1). (B) A sequence alignment of various type 2 CsoR operator sites identified in S. lividans [Sli (11)], Staphylococcus aureus [Sau (12)], Listeria monocytogenes [Lmo (33)] and B. subtilis [Bsu (8)]. The presence of a 5′-TAC/GTA-3′ inverted repeat is noted as a consistent element in the alignment consensus, with the M. tuberculosis binding site having an inverse orientation of this inverted repeat. The approximate lengths of various DNA constructs of csoR-EXT used as probes to study the binding characteristics of CsoRSl are indicated, highlighting similar dimensions to the tetrameric CsoRSl width with that of the 5′-TAC/GTA-3′ inverted repeat. (C) A proposed schematic of the CsoR symmetry on binding to its operator, with the region of the GTA dyads shown as grey regions on the DNA.