Fig. 1.
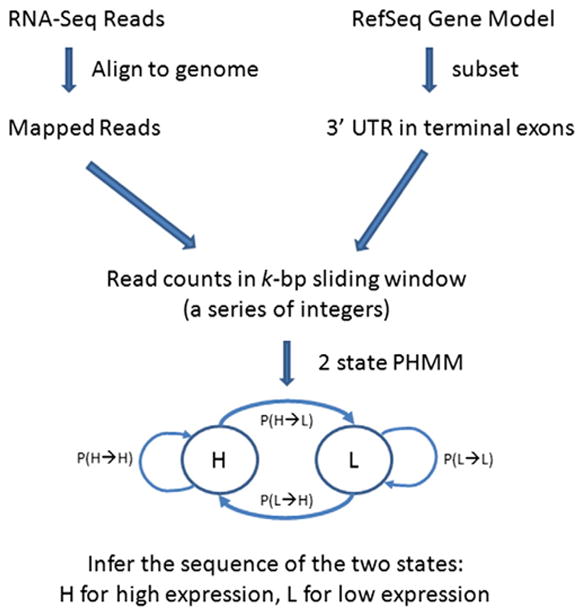
Workflow to model shortened polyA sites. A Poisson hidden Markov model (PHMM) is constructed to infer the sequence of two states using a series of integers obtained from RNA-Seq reads mapped to the 3′ untranslated region (UTR) of RefSeq transcripts. The two states are H for high expression and L for low expression. The lines with the arrows represent transitions from state to state. P (X→Y) is the probability of transitioning from state X to state Y. Note that we only selected transcripts with transitions from H→L in detecting APA.
