Fig. 4.
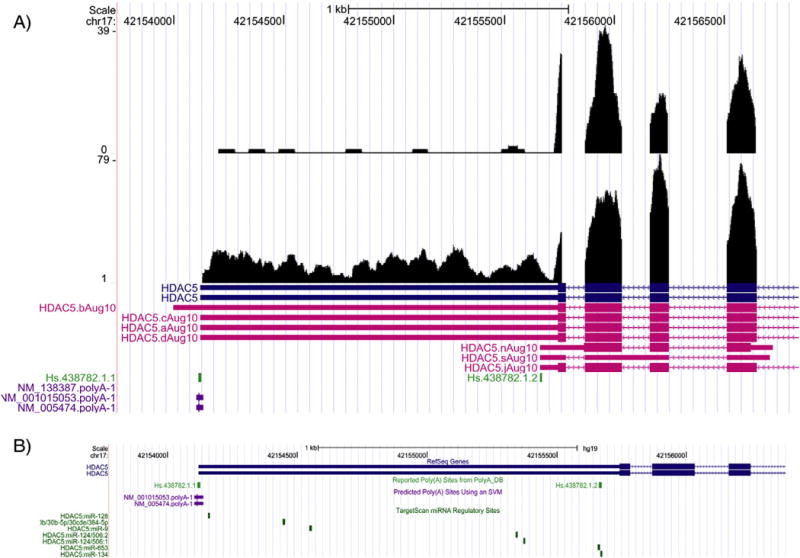
Differential alternative polyadenylation of the HDAC5 gene. A) The HDAC5 gene, shown on the antisense strand of chromosome (chr) 17, is represented by RefSeq accession# NM_005474 and UniGene cluster IDs Hs.438782.1.1 and Hs.438782.1.2 and contains the mapping of RNA-Seq reads at the 3′ UTR in liver (top track) and cortex (bottom track) tissues. The predicted polyA sites (NM_138387.polyA-1, NM_001015053.polyA-1, and NM_005474.polyA-1) are shown. The x-axis denotes the chr position and the y-axis reflects the coverage of the mapped reads. B) Shown are the potential targets of 7 miRNAs (from the TargetScan database) to the 3′ UTR of the HDAC5 gene. The x-axis denotes the chr position.
