Fig. 1.
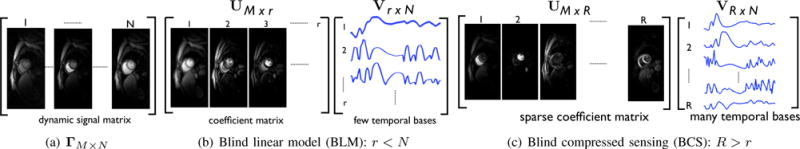
Comparison of blind compressed sensing (BCS) and blind linear model (BLM) representations of dynamic imaging data: The Casorati form of the dynamic signal Γ is shown in (a). The BLM and BCS decompositions of Γ are respectively shown in (b) and (c). BCS uses a large over-complete dictionary, unlike the orthogonal dictionary with few basis functions in BLM; (R > r). Note that the coefficients/spatial weights in BCS are sparser than that of BLM. The temporal basis functions in the BCS dictionary are representative of specific regions, since they are not constrained to be orthogonal. For example, the 1st, 2nd columns of UM×R in BCS correspond respectively to the temporal dynamics of the right and left ventricles in this myocardial perfusion data with motion. We observe that only 4-5 coefficients per pixel are sufficient to represent the dataset.
