Figure 2.
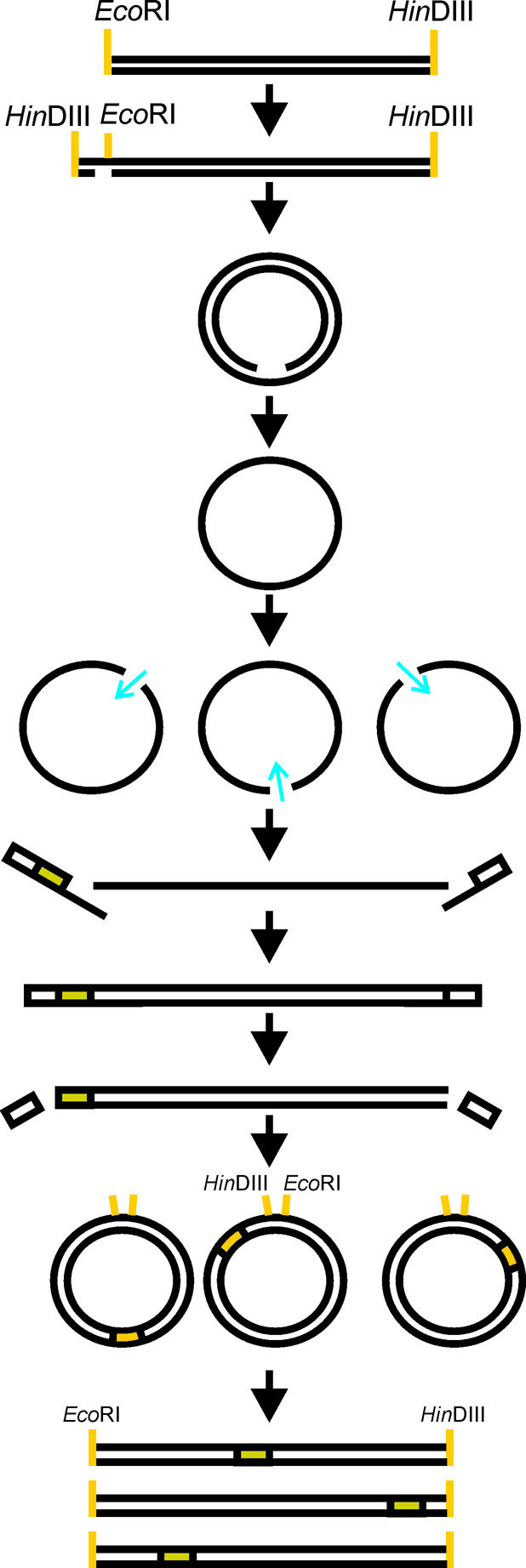
Random insertion/deletion mutagenesis (RID). The template DNA is converted to a covalently closed single-stranded circle which is cleaved at random sites by Ce(IV)-EDTA treatment. Linker fragment cassettes are then annealed to each end of the cleaved single-stranded DNA via a 10 nt random tail. The construct is amplified using primer sites in the cassettes to produce the second DNA strand. Finally the cassettes are cleaved off using a type II restriction enzyme (recognition site in the cassette) to leave the insertion or deletion behind. The remaining construct containing the modification is converted back to double-stranded circular DNA that can be cleaved with appropriate restriction sites to produce the gene library in a form ready for cloning. Adapted from Murakami et al. (26) with permission from Nature Publishing Group (http://www.nature.com/nbt/).
