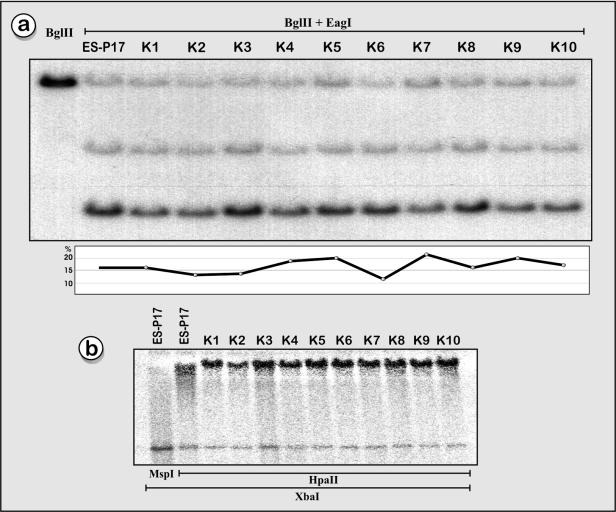
Figure 3.
Stability of DNA methylation in imprinted neuronal genes in single cell-derived clonal ES cell lines. (a) Methylation-sensitive Southern blot analyses with restriction enzymes BglII and EagI revealed only small variations in methylation patterns of the Ndn gene in 10 Bruce 4 (C57BL/6) ES cell clones (Ndn probe a). (Bottom) The relative methylation levels in all ES cell subclones (K1–K10) varied around 15% (±5%) and were nearly identical to the methylation level observed in the original cell line (ES-P17). (b) Phosphoimager scan of a Southern blot with genomic DNA of 10 Bruce 4 subclones (5 µg/lane), cleaved with XbaI and the methylation-sensitive HpaII. The DNA on the membrane was hybridized with a mouse Snurf/Snrpn DMR1 probe. The first lane represents a control with the HpaII-isoschizomer MspI, which cleaves DNA also at methylated 5′-CCGG-3′ sites. The Snurf/Snrpn DMR1 was hypermethylated (65 ± 5%) in the Bruce 4 cell line ES-P17 and in all 10 subclones derived from these ES cells.