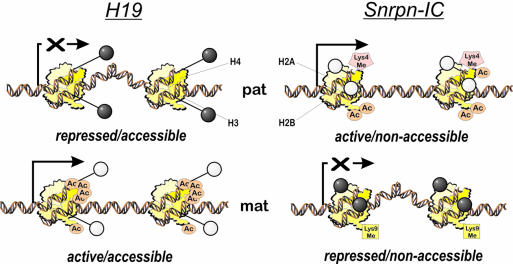
Figure 7.
The Snrpn IC and the H19/Igf2 imprinting region may be differentially accessible to modifying enzymes. It seems possible that the overall chromatin structure in these regions may contain several dynamic levels of regulation. For example, a DNA sequence could be either in a methylated repressed state (heterochromatin) or in an active unmethylated conformation (euchromatin). Independent of these conformations, modifying enzymes could have different access to these chromosomal regions. Since the Snrpn IC and H19/Igf2 region are active in ES cells, they could both have an open chromatin structure on their active alleles, but may expose their CpG dinucleotides differently due to a different nucleosomal structure. The H19 region was reported to be accessible to methyltransferases in ES cells (50), which could lead to an increased susceptibility to epigenetic changes during in vitro culture. In contrast, the Snrpn IC could be already in an epigenetic maintenance mode, independent of its transcriptional activity. Both genomic regions have complex patterns of known epigenetic modifications (49,51,75–77). Histone H3 on the paternal allele has lysine 4 methylation and is acetylated, accompanied with a hypomethylation of the CpG dinucleotides. On the other hand, on the maternally inherited allele, chromatin is marked by hypermethylation on lysine 9 of H3 and hypermethylation of the DNA (75). Dark lollipops, CpG methylated; white lollipops, CpGs are unmethylated; Ac, histone-acetylation; Me, histone methylation; Lys, lysine residues.