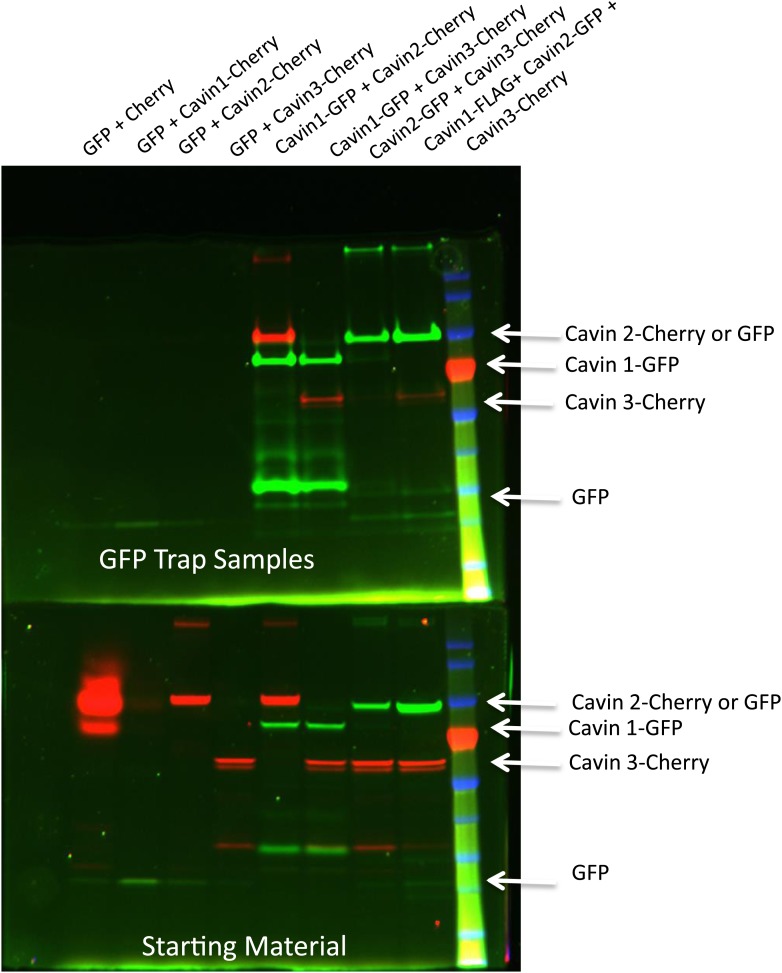
Figure 2. Single-molecule coincidence and ALPHAScreen mapping of interactions among cavin1, 2 and 3.
(A) Histogram of single-molecule coincidence between cavin1-GFP and cavin2-Cherry co-transfected in MCF-7 cells. The coincidence is calculated as the ratio of intensity in the Cherry channel divided by the sum of the signals in the GFP and Cherry channels. GFP-only bursts show a coincidence at 0 and Cherry-only oligomers are located at coincidence = 1. For the oligomers containing both fluorophores, the coincidence ratio is a measure of the stoichiometry of the assembly. (B) Same as (A), coincidence between cavin1-GFP and cavin3-Cherry. (C) Same as (A), coincidence between cavin2-GFP and cavin3-Cherry. (D–F): using cell-free protein expression, three-dimensional histograms of single-molecule coincidence between GFP and Cherry cavin proteins at expression ratios spaning from 100% GFP to 100% Cherry. For each DNA ratio of cavin-GFP and cavin-Cherry, we collected >1000 bursts and plotted the corresponding histograms of coincidence. We varied the ratio of cavins and created a stack of 10 histograms representing the various stoichiometries. The histograms for individual pairs were then aggregated into 3D plots. (D) Evolution of mixed oligomers of cavin1-GFP and cavin2-Cherry revealing formation of oligomers with a full range of stoichiometries. (E) The cavin1 and cavin3 plot shows formation of cavin1-cavin3 oligomers with predominantly 3/1 composition. (F) The cavin2 and cavin3 plot shows that these proteins do not form mixed oligomers. (G) Schematic representation of ALPHAScreen principle. This bead–bead assay relies on transfer of singlet oxygen from a donor bead to a luminescent acceptor bead when protein–protein interactions bring the beads within 200 nm (see ‘Materials and methods’ and SI for details). (H) A plot of ALPHAScreen signal across the concentrations of cavin proteins attached to donor and acceptor beads. The interactions for cavin2 and cavin3 in the presence or absence of cavin1 display amplitudes close to the background signal. (I) Values obtained for cavin1-GFP and cavin2-myc, cavin1-GFP and cavin3-myc, cavin2-GFP and cavin2-myc reveal robust interactions. However the curves obtained for cavin3-GFP and cavin2-myc, cavin2-GFP and cavin3-myc demonstrate that cavin2 and cavin3 cannot bind to each other. The triple co-expression of cavin2-GFP, cavin3-myc and untagged cavin1 results in no change in binding, suggesting that cavin1 cannot act as a bridge between cavin2 and cavin3.