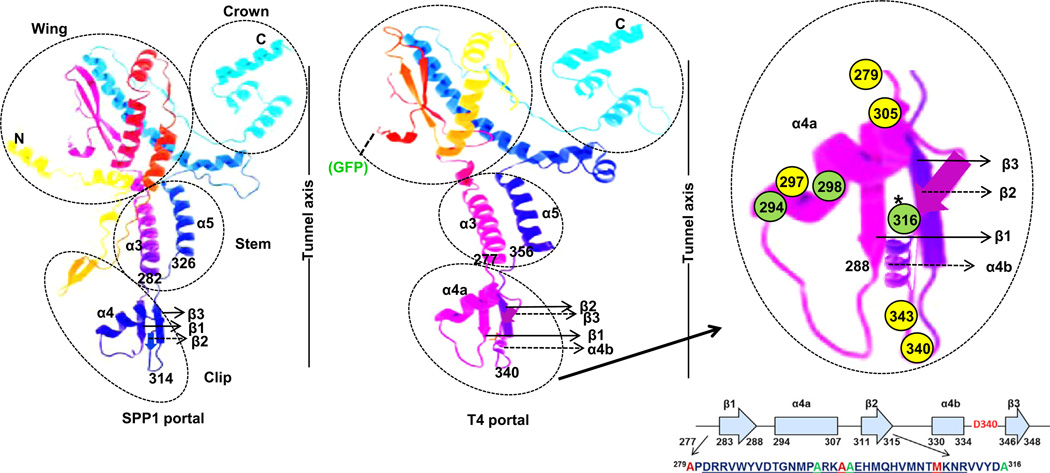
Fig. 5.
Tertiary structure prediction of T4 portal using the Phyre2 program. Model is based on template (PDBID-2jes) (SPP1 portal). 296 residues (56% of sequence) have been modeled with 78.7% confidence by the single highest scoring template. Of amino acids assigned in the clip region, those in yellow circles could not be substituted by cysteine (279, 297 and 305) or with any other amino acids (340, 343), amino acids in green circles could be substituted with cysteine. FRET transfer between CT-ReAsH terminase and Alexa 488 labeled proheads was observed with A316C residue modification. Secondary structures that are predicted by the Phyre2 program are shown by solid arrows whereas those predicted by PSIPRED analysis and not by the Phyre2 program are shown by dashed arrows. The Crown, Wing, Stem, and Clip regions of the SPP1 portal crystal structure are shown in relation to the DNA tunnel axis. The position of the N-terminal GFP in the predicted T4 portal is shown.