Abstract
Tight control of antigen-receptor gene rearrangement is required to preserve genome integrity and prevent the occurrence of leukemia and lymphoma. Nonetheless, mistakes can happen, leading to the generation of aberrant rearrangements, such as Tcra/d-Igh inter-locus translocations that are a hallmark of ATM deficiency. Current evidence indicates that these translocations arise from the persistence of unrepaired breaks converging at different stages of thymocyte differentiation. Here we show that a defect in feedback control of RAG2 activity gives rise to bi-locus breaks and damage on Tcra/d and Igh in the same T cell at the same developmental stage, which provides a direct mechanism for generating these inter-locus rearrangements. Both the RAG2 C-terminus and ATM prevent bi-locus RAG-mediated cleavage through modulation of 3D conformation (higher order loops) and nuclear organization of the two loci. This limits the number of potential substrates for translocation and provides an important mechanism for protecting genome stability.
B and T lymphocyte development is driven by V(D)J recombination, a process through which V, D and J coding segments within each of the seven antigen receptor loci, are rearranged to create a vast repertoire of antigen receptors1,2. Generation of receptor diversity through recombination is critical for shaping the adaptive arm of the immune system, enabling B and T cells to mount a focused and specific response to foreign antigen. This programmed rearrangement event relies on the lymphoid-specific proteins, RAG1 and RAG2 (Recombination Activating Genes 1 and 2), which individually harbor many distinct regulatory domains whose functions remain largely enigmatic. Nonetheless, it is known that at least some of these contribute to the performance of RAG through fine-tuning of targeting, cleavage and repair. Furthermore, the proper functioning of the recombinase complex relies on cooperation between the two proteins, RAG1 and 2.
Specificity of targeting is conferred by RAG1 mediated recognition of highly conserved recombination signal sequence (RSS) elements that flank the individual V, D and J gene coding segments, which are arrayed along each antigen receptor locus3-5. Moreover, RAG1 carries the catalytic endonuclease activity4,5. However, cleavage cannot occur in the absence of its partner protein, RAG26,7, which contains a PHD domain (Plant Homeo Domain) that is known to direct binding of the recombinase to active chromatin through recognition of the histone modification, H3K4me38,9. The RAG1/2 complex binds to two gene segments (that can be many kilobases apart) brings them together and cuts at the RSS borders to generate DNA double-strand breaks (DSBs). Following cleavage the four resulting broken ends are held together in a RAG post-cleavage complex which is instrumental in directing repair by the ubiquitous non-homologous end joining (NHEJ) pathway1,2,10,11. The introduction of DSBs activates several PI3K-like Ser/Thr kinases, including the ATM kinase (Ataxia telangiectasia mutated), which phosphorylate downstream proteins and orchestrate the DNA damage response2. Other DNA damage response factors, like the histone variant γ-H2AX, 53BP1 (p53 binding protein 1), and the MRN complex (containing Mre11, Rad50 and Nbs1), are rapidly recruited and form nuclear foci at the site of DSBs2,11.
Recombination is tightly regulated so that the appropriate loci and gene segments are rearranged in the appropriate lineage (T cell receptor (Tcr) loci in T cells and immunoglobulin (Ig) loci in B cells) and at the appropriate developmental stage. In T cells productive rearrangement of the different Tcr loci gives rise to two distinct lineages: recombination of Tcrg/Tcrd and Tcrb/Tcra leads to γδ and αβ T cells, respectively12,13. Despite this separation, recombination of the different loci overlaps. Tcrg, Tcrd and Tcrb are all rearranged at the early CD4−CD8− double negative DN2/3 stage of development, while Tcra recombination occurs later in double positive (DP) cells14.
Regulation of Tcrd and Tcra recombination is uniquely complicated because, beyond the fact that they recombine at different stages of differentiation, Tcra and Tcrd share the same chromosomal location, with the latter embedded between the Vα and Jα gene segments. Furthermore, promiscuous DH-to JH rearrangement of the Igh locus, which occurs at low level in T lineage cells15, adds yet another layer of complexity. Together these issues compound the risks associated with Tcra/d recombination and the probability of aberrant repair. Indeed, inter-locus rearrangements between Tcra/d and Igh have been identified as a hallmark of thymic lymphomas in ATM-deficient mice16. Moreover, we recently discovered translocations between these two loci associated with an absence of the non-core C-terminal domain of RAG217. Although this domain is dispensable for recombination18,19, its deletion is known to affect the joining step, as well as the order, efficiency and fidelity of the reaction10,20-25. When coupled with the disruption of p53, we found that Rag2c/c p53−/− mice develop thymic lymphomas harboring recurrent translocations involving Tcra/d and Igh, defects that are similar to those found in Atm−/− mice17. In addition, ATM and the C-terminus of RAG2 have similar defects in stabilizing the RAG post-cleavage complex17,26.
Here we have now investigated the mechanisms underlying the origins of the inter-locus Igh-Tcra/d translocations prior to lymphomagenesis to determine whether regulation of cleavage and nuclear accessibility of the loci is perturbed by an absence of ATM and the C-terminus of RAG2. We find that Igh cleavage occurs at higher levels in DN2/3 versus DP cells and thus its rearrangement could overlap with Tcra/d rearrangement. However breaks are not found in Igh and Tcra/d in the same cell, except in the absence of the RAG2 C-terminus or ATM. Control of mono-locus cleavage involves regulated mono-locus looping out from the chromosome territory and mono-locus association with repressive pericentromeric heterochromatin. In the absence of the RAG2 C-terminus or ATM nuclear accessibility is increased and both loci remain euchromatic and bi-locus loops can be detected coincident with bi-locus cleavage. Interestingly we found that expression of RAG brings Tcra/d and Igh into close proximity in DN2/3 cells (when RAG-mediated cleavage Igh occurs at high levels) while the two loci separate at the subsequent DP cell stage of development (when recombination of Igh is reduced). In contrast, in the mutant cells increased association of Tcra/d-Igh in DP cells is linked to increased nuclear accessibility, and the introduction of bi-locus breaks and damage on proximal loci. In sum, this study show a role for the non-core domain of RAG2 and of ATM in controlling recombination between two loci via modulation of nuclear organization. Moreover, these events provide a direct mechanism for the generation of the Tcra/d-Igh translocations that are found in Rag2c/c p53−/− and Atm−/− tumors.
Results
Igh and Tcra/d rearrangement overlaps in DN2/3 and DP cells
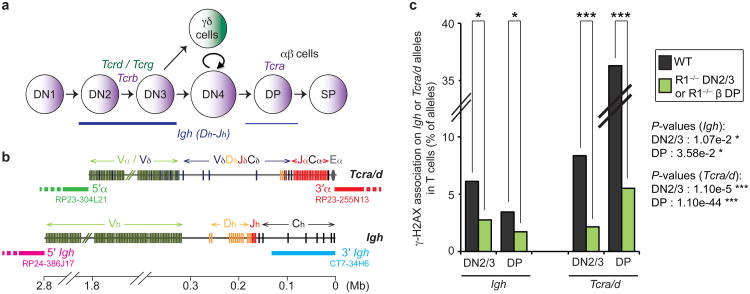
Although it is established that the Igh locus (which is located on chromosome 12) undergoes low level DH-JH rearrangement (at the 3′ end of the locus) in T cells, it is not known at which stage of development recombination occurs. To determine this, we performed an immuno-DNA FISH experiment to analyze RAG mediated cleavage on Igh in wild-type DN2/3 and DP cells (Fig. 1a) using a DNA probe that hybridizes to the 3′ end of this locus (See scheme Fig. 1b) in combination with an antibody against the phosphorylated form of H2AX, γ-H2AX, as a read-out for double-stranded breaks (DSBs)27,28. Our analyses showed that RAG-mediated γ-H2AX foci were associated more frequently with Igh in DN2/3 compared to DP cells suggesting that DH-JH rearrangement occurs predominantly, but not exclusively, at the earlier stage of development (Fig. 1c and Supplementary Table S1). In contrast, RAG-mediated γ-H2AX foci were associated more frequently with Tcra/d in DP compared to DN2/3 cells (Fig. 1b,c and Supplementary Table S1).
Fig. 1.
Igh and Tcra/d rearrangement overlaps in DN2/3 and DP T cells. (a) Scheme of T cell development with stages of recombination of the 4 Tcr loci. (b) Scheme representing the Tcra/d and Igh loci with probes for 3D-DNA FISH shown below. (c) Frequency of γ-H2AX association on Igh or Tcra/d alleles in WT DN2/3 and DP cells. P-values were calculated using a two-tail Fisher exact test (-ns- no significance (P ≥ 5.00e-2), -*- significant (5.00e-2 > P ≥ 1.00e-2), -**- very significant (1.00e-2 > P ≥ 1.00e-3), -***- highly significant (P < 1.00e-3)). Experiments were repeated at least two times and data are displayed as a combination of two independent experimental sets (n > 500 for each stage/genotype; See Supplementary Table S1 for details and individual data sets).
The RAG2 C-terminus and ATM regulate mono-locus cleavage
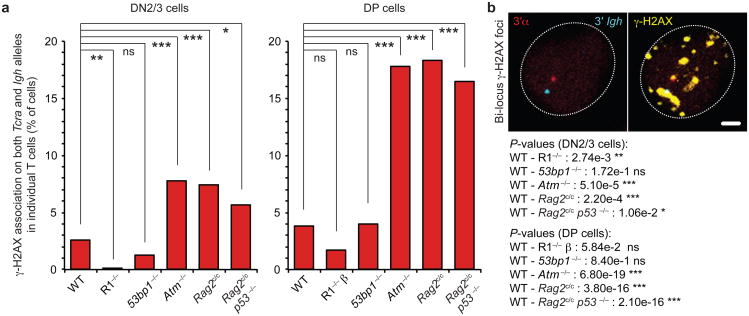
As Tcra/d and Igh loci both undergo recombination in T cells, it is conceivable that there is overlap in the timing of their rearrangement. If so, bi-locus breaks could be introduced concurrently in the same cell and in the event of a defect in joining, these could act as substrates for translocations. However, when we examined the frequency of γ-H2AX foci associated with both Tcra/d and Igh (bi-locus breaks) in individual wild-type DN2/3 and DP cells we found these present at a very low level which was only slightly above the frequency of bi-locus breaks detected in RAG-deficient cells (Fig. 2a and Supplementary Tables S2, S3). In contrast, we found a significant rise in the frequency of bi-locus breaks in individual Rag2c/c p53−/− or Atm−/− cells, with the most pronounced increase occurring at the DP stage, at the time of Tcra recombination (Fig. 2a,b and Supplementary Tables S2, S3). It is of note that we found no increase in the frequency of bi-locus breaks in the absence of another DNA damage response factor, 53BP1, which is known to be important for the long-range joining of coding ends in V(D)J recombination29 (Fig. 2a and Supplementary Tables S2, S3). These data indicate that bi-locus breaks in the same cell do not occur simply as a result of a defect in repair.
Fig. 2. The RAG2 C-terminus and ATM regulate mono-locus cleavage.
(a) Frequency of γ-H2AX association on both Tcra/d and Igh loci in individual WT, Rag1−/− (R1−/− or R1−/− β (See Methods section for details)), 53bp1−/−, Atm−/−, Rag2c/c and Rag2c/c p53−/− DN2/3 and DP cells. (b) Confocal sections showing a representative example of γ-H2AX association (in yellow) on both Tcra/d and Igh loci (3′α in red, 3′Igh in blue). Scale bar = 1 μm. P-values were calculated using a two-tail Fisher exact test (-ns- no significance (P ≥ 5.00e-2), -*- significant (5.00e-2 > P ≥ 1.00e-2), -**- very significant (1.00e-2 > P ≥ 1.00e-3), -***- highly significant (P < 1.00e-3)). Experiments were repeated at least two times and data are displayed as a combination of two independent experimental sets (n > 200 for each stage/genotype; See Supplementary Tables S2, S3 for details and individual data sets).
Since cells deficient in both p53 and ATM have defects in cell cycle checkpoints that enable unrepaired breaks to be propagated as a result of cell division30,31 it is possible that some proportion of the bi-locus breaks that we detected in mutant DP cells could be attributed to the persistence of unrepaired breaks being amplified in proliferating DN4 cells, which mark the transition from the DN2/3 to the DP stage of development (Fig. 1a). To check this we compared the frequency of bi-locus breaks in individual Rag2c/c versus Rag2c/c p53−/− cells. However, we found no significant differences in the incidence of bi-locus breaks in the two genotypes so we conclude that the increase in bi-locus breaks that we detected in Rag2c/c p53−/− DP cells does not result from an absence of p53 (Fig. 2a and Supplementary Tables S2, S3). Indeed, we also found no significant increase in bi-locus breaks on Tcra/d and Igh in wild-type versus p53−/− cells (4% compared to 4.6%, respectively). In addition, differences in the level of ATM in Rag2c/c versus wild-type cells cannot explain the differences in the frequency of bi-locus breaks as Western blot analysis showed similar levels of protein in the two genotypes (Supplementary Fig. S1). Taken together these data indicate that the C-terminus of RAG2 and ATM regulate cleavage to ensure that RAG mediated breaks are introduced on only one locus at a time in each recombining T cell.
The RAG2 C-terminus and ATM regulate Tcra-Igh association
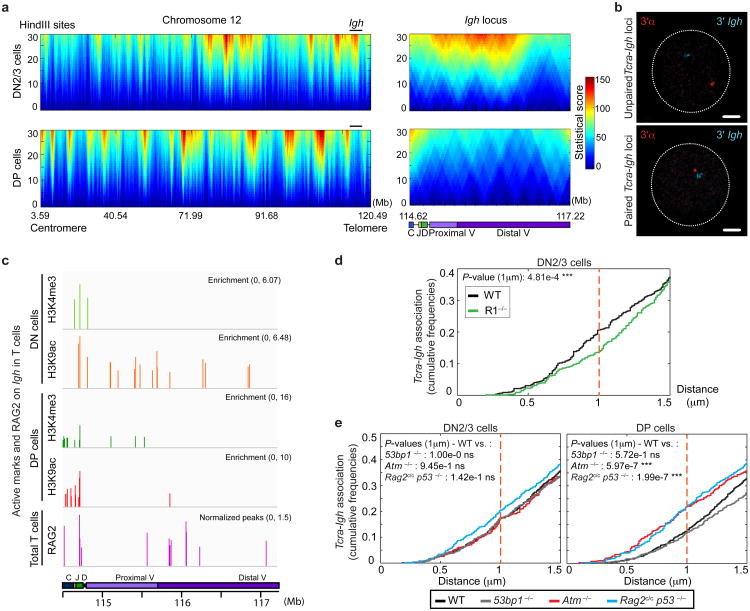
We and others have shown that nuclear proximity of broken partner genes is an important factor in translocations32-37. Since our studies indicate that translocations between Tcra/d and Igh are found in tumors from Rag2c/c p53−/− and Atm−/− mice17 we wanted to examine the association between the two loci in recombining DN2/3 and DP cells. For this we performed circularized chromosome conformation capture with next generation sequencing (4C-seq), using a bait sequence spanning the Tcra enhancer (Eα) at the 3′α end of the locus (see Methods section for details; Fig. 3a, Supplementary Fig. S2a and Supplementary Table S4). The 4C data is displayed as a domainogram, which is a statistical way to visualize local 4C signal enrichment across a region of interest38. It is clear from Fig. 3a (left panels) that there are significant changes in the interaction partners of Tcra/d in the two populations across chromosome 12. Importantly, the intensity of the Tcra/d-Igh interaction is much higher in DN2/3 cells where, according to our γ-H2AX analysis, the latter is recombined at higher frequency (Fig. 3a (right panels)). This result was validated by additional 3-D DNA FISH experiments, and distances separating the two loci were plotted as cumulative frequency curves, using a cut-off of 1μ m to measure close association (ie. ‘pairing’) (Supplementary Fig. S2b). On these graphs a left shift is indicative of closer association. Examples of paired and unpaired loci are shown in Fig. 3b.
Fig. 3. The RAG2 C-terminus and ATM regulate Tcra-Igh association.
(a) Domainograms showing a heatmap of Tcra/d interactions across the entire chromosome 12 (left) and the Igh locus (right) using a window size of 30 HindIII sites in DN2/3 (top) and DP (bottom) cells. See also Supplementary Fig. S2 and Table S4. (b) Confocal sections showing representative examples of unpaired (top) and paired (bottom) loci. 3′α in red, 3′Igh in blue. Scale bars = 1 μm. (c) Alignment of ChIP-seq data at the Igh locus showing levels of enrichment of H3K4me3 (green), H3K9ac (red) and RAG2 binding (purple)39 in DN and DP cells. See also Supplementary Table S5. (d,e) Cumulative frequency curves of Tcra-Igh inter-locus distances in: WT and R1−/− DN2/3 cells (d), WT, 53bp1−/−, Atm−/− and Rag2c/c p53−/− DN2/3 (e, left) and DP (e, right) cells (cut-off at 1.5 μm). A left shift indicates closer association. P-values were calculated using a two-tail Fisher exact test (-ns- no significance (P ≥ 5.00e-2), -*- significant (5.00e-2 > P ≥ 1.00e-2), -**- very significant (1.00e-2 > P ≥ 1.00e-3), -***- highly significant (P < 1.00e-3)). Experiments were repeated at least two times and data are displayed as a combination of two independent experimental sets (n > 200 for each stage/genotype).
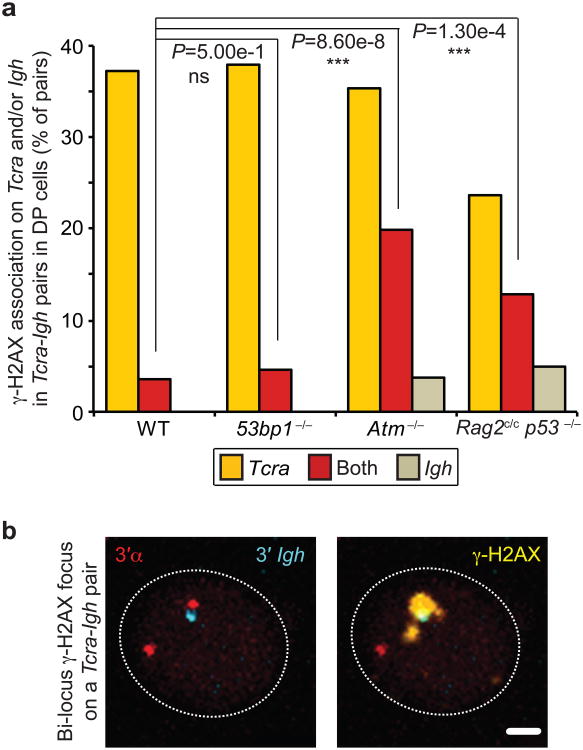
Previous studies indicate that RAG binds to active chromatin and localizes to the J segments at the 3′ end of each antigen receptor locus in rearranging cells39. We have recently shown that localized RAG enrichment in this region is linked with homologous Tcra pairing, transcription and regulated mono-allelic cleavage in DP cells40. Thus, we wanted to investigate whether the presence of RAG could influence the frequency with which Tcra/d and Igh contact each other during recombination. There are two regions of RAG2 binding on the Igh locus in developing T cells (Fig. 3c). The first corresponds to enrichment of H3K4me3 and H3K9ac at the 3′ end of Igh, where DH-JH rearrangement is known to occur in these cells15. In addition, RAG2 also binds to the VH portion of Igh and this could explain the high frequency of interaction with Tcra in this region (Fig 3a and Supplementary Table S5). Interestingly, we found that an absence of RAG1 significantly decreased heterologous association of the two loci in DN2/3 cells while we observed no significant change in DP cells (Fig. 3d and Supplementary Fig. S2c). These data indicate that RAG1 brings recombining Tcra/d and Igh together in DN2/3 cells (where Igh cleavage occurs at high levels) but that these loci separate in DP cells (where cleavage of Igh is reduced and cleavage of Tcra occurs at high levels) (Fig. 1c). Intriguingly, the absence of the C-terminus of RAG2 or ATM deficiency did not affect the frequency of Tcra/d-Igh association in DN2/3 cells, however in DP cells, where association of the two loci is normally reduced, we observed an increase in the incidence of heterologous pairing in both mutants, while pairing in the equivalent 53BP1-deficient populations followed the same pattern as in wild-type cells (Fig. 3e). Importantly, bi-locus breaks were increased on heterologously paired loci in Rag2c/c p53−/− and Atm−/− DP cells compared with wild-type control and 53BP1-deficient cells (Fig. 4a,b and Supplementary Table S6). In sum, these data indicate that an increased frequency in contact between Tcra and Igh in Rag2c/c p53−/− and Atm−/− DP cells is linked to an increased frequency in bi-locus breaks on paired loci.
Fig. 4. RAG2 C-terminus and ATM control cleavage on Tcra-Igh pairs.
(a) Frequency of γ-H2AX association on Tcra and/or Igh in Tcra-Igh pairs in WT, 53bp1−/−, Atm−/−and Rag2c/c p53−/− DP cells. (b) Confocal sections showing a representative example of Tcra-Igh pair with a γ-H2AX focus on both loci. 3′α in red, 3′Igh in blue and γ-H2AX in yellow. Scale bar = 1 μm. P-values were calculated using a two-tail Fisher exact test (-ns- no significance (P ≥ 5.00e-2), -*- significant (5.00e-2 > P ≥ 1.00e-2), -**- very significant (1.00e-2 > P ≥ 1.00e-3), -***- highly significant (P < 1.00e-3)). Experiments were repeated at least two times and data are displayed as a combination of two independent experimental sets (n > 80 for each genotype; See Supplementary Table S6 for details and individual data sets).
RAG-mediated pairing is not a general phenomenon
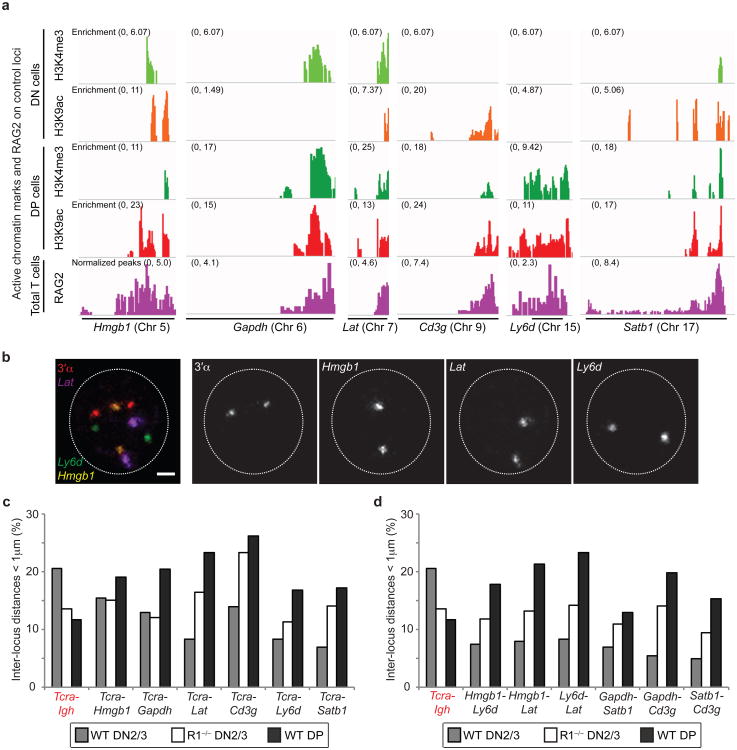
Genome wide analysis has shown that RAG2 has an overlapping pattern with H3K4me3 enriched regions39, thus we wanted to determine whether other RAG enriched active genes would pair up with Tcra in the same way as Igh. For this analysis we selected five hematopoietic lineage specific genes (Hmgb1, Lat, Cd3g, Ly6d and Satb1) as well as a housekeeping gene (Gapdh) that are all located on different chromosomes and transcribed in these cells (Fig.5a,b). Interestingly, pairing of Tcra with these other RAG-enriched loci shows an opposite trend to Igh-Tcra pairing in all cases (Fig. 5c and Supplementary Table S7). Indeed, in contrast to Tcra-Igh an absence of RAG1 appears to increase their contact frequency with Tcra in DN2/3 cells and association is even more pronounced at the DP stage of development. Furthermore, pairing between these control loci follows the same trend as their association with Tcra (Fig. 5d and Supplementary Table S7). It is of note that the maximum frequency of pairing (for individual pairs of alleles) in all cases is very similar (around 20%) and this matches the maximum level of homologous Tcra pairing seen during recombination in DP cells40. Taken together these data indicate that the trend for Tcra-Igh is unique and even though RAG2 is enriched on control loci, pairing does not depend on the presence of RAG. Instead, interactions between control loci may be influenced by binding of other transcription factors that are involved in their regulation. In this context the Fraser lab have shown that co-ordinately regulated genes associate with each other in common transcription factories and more recent studies from the Murre lab indicate that transcription factor bound regions determine intra- and inter-domain interactions that are developmentally regulated41,42. Thus, perhaps other transcription factors that are enriched on these control genes may be dominant in determining their interaction partners in developing T cell nuclei.
Fig. 5. RAG-mediated pairing is not a general phenomenon.
(a) Alignment of ChIP-seq data at the Hmgb1, Gapdh, Lat, Cd3g, Ly6d and Satb1 loci showing levels of enrichment of H3K4me3 (green), H3K9ac (red) and RAG2 binding (purple)39 in DN and DP cells. (b) Confocal sections showing 3D DNA FISH for Tcra and three control loci (3′α in red, Hmgb1 in yellow, Lat in purple and Ly6d in green. Scale bar = 1 μm. (c,d) Graphs showing the frequency of heterologous pairing (inter-locus distance < 1μm) between Tcra and the control loci (c), or between the loci (d). Experiments were performed at least one time and data are displayed as a combination of independent experimental sets when applicable (n > 200 for each stage/genotype; See Supplementary Table S7 for details).
Regulation of RAG cleavage is linked to genome stability
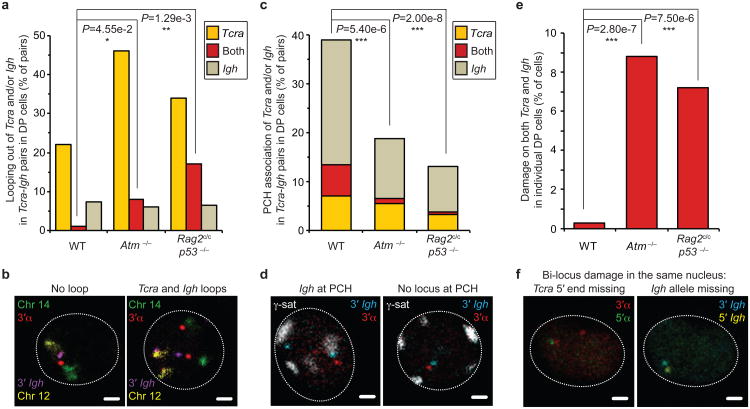
The Tcra and Igh loci are located on different chromosomes, namely chromosomes 14 and 12 respectively. To understand how association between these two loci occurs in nuclear space we focused our attention on the formation of higher order loops which moves genes outside of their chromosome territories as a mechanism by which they could be brought into close contact. We recently showed that mono-locus looping out of Tca is linked to homologous pairing during recombination40. To examine this we measured the distance separating the 3′ end of Tcra and Igh from their respective chromosome 14 and 12 territories when the loci were paired. Our analyses indicate that in wild-type DP cells the vast majority of higher-order loop formation occurred on one locus at a time and these predominantly involved Tcra (Fig. 6a,b and Supplementary Table S8), the locus in which we detected the most breaks (Fig. 4a). In contrast, there was a significant increase in bi-locus loop formation on paired Tcra-Igh alleles in Rag2c/c p53−/− and Atm−/− DP cells (Fig. 6a,b and Supplementary Table S8) coincident with an increase in bi-locus breaks (Fig. 4a). Taken together our data indicate that an increase in Tcra-Igh pairing in Rag2c/c p53−/− and Atm−/− DP cells is linked to an increase in bi-locus looping and bi-locus breaks on paired alleles.
Fig. 6. Regulation of RAG cleavage is linked to genome stability.
(a) Frequency of higher-order looping out of the 3′ ends of Tcra and/or Igh in Tcra-Igh pairs in WT, Atm−/−and Rag2c/c p53−/− DP cells. (b) Confocal sections showing examples of Tcra-Igh pairs with no loop or looping of both Tcra and Igh. 3′α in red, 3′Igh in purple, chromosome 14 in green and 12 in yellow. Scale bars = 1 μm. (c) Frequency of PCH association of Tcra and/or Igh in Tcra-Igh pairs in WT, Atm−/−and Rag2c/c p53−/− DP cells. (d) Confocal sections showing representative examples of Tcra-Igh pairs with Igh located at PCH or no locus at PCH. 3′α in red, 3′Igh in blue and γ-satellite (PCH) in white. Scale bars = 1 μm. (e) Frequency of cells with bi-locus damage on both Tcra/d and Igh alleles in individual WT, Atm−/−and Rag2c/c p53−/− DP cells. (f) Confocal sections showing a representative example of bi-locus damage on both Tcra and Igh alleles in the same nucleus. One Tcra 5′ end and one Igh allele are missing. 3′α in red, 5′α in green, 3′Igh in blue and 5′Igh in yellow. Scale bars = 1 μm. P-values were calculated using a two-tail Fisher exact test (-ns- no significance (P ≥ 5.00e-2), -*- significant (5.00e-2 > P ≥ 1.00e-2), -**- very significant (1.00e-2 > P ≥ 1.00e-3), -***- highly significant (P < 1.00e-3)). Experiments were repeated at least two times and data are displayed as a combination of two independent experimental sets (n > 50 for each genotype; See Supplementary Tables S8-S10 for details).
Higher-order looping of genes away from their chromosome territories has previously been correlated with open chromatin and an active transcriptional status, while silent genes are positioned more internally43-46. To determine whether nuclear accessibility of the individual loci is linked to loop formation we examined the location of paired Tcra-Igh relative to repressive pericentromeric heterochromatin (PCH). In wild-type DP cells we found that paired Tcra-Igh alleles were frequently located at PCH, however the two loci were not equivalently close (Fig. 6c,d and Supplementary Table S9). Interestingly, we found that the Igh locus was predominantly in contact with PCH while Tcra, the locus associated with the most loops and breaks in these pairs, remained euchromatic. In contrast, in Rag2c/c p53−/− and Atm−/− DP cells, repositioning of the Igh to PCH was significantly reduced and this increased nuclear accessibility was associated with increased bi-locus breaks (Fig. 6c,d). In sum, these data indicate that the increase in bi-locus breaks on paired Tcra-Igh alleles in Rag2c/c p53−/− and Atm−/− DP cells is linked with an increase in the simultaneous nuclear accessibility of the two loci, as assessed by looping out from their chromosome territories and by their location away from repressive PCH.
Since close proximity of broken partner loci in DP cells could provide a fertile ground for the generation of the aberrant inter-locus rearrangements that are a characteristic of Rag2c/c p53−/− and Atm−/− lymphomas we next asked whether the increase in bi-locus breaks and increased accessibility of paired Tcra/d/Igh was linked with an increase in bi-locus instability in interphase Rag2c/c p53−/− and Atm−/− cells prior to the onset of lymphomagenesis. For this, we performed a DNA FISH experiment using BAC probes that hybridize outside the 3′ and 5′ ends of each locus (See scheme Fig. 1b) and examined the frequency of damage (ie. split alleles (> 1.5μm in between the two ends), duplicated or missing signals). The increase in bi-locus damage in individual Rag2c/c p53−/− and Atm−/− mutant DP cells was very significant (Fig. 6e,f and Supplementary Table S10). Thus an absence of the C-terminal domain of RAG2 or ATM results in an increase in both bi-locus breaks and bi-locus damage in the same cell.
Discussion
Taken together our data show that translocations between Tcra/d and Igh that are found in tumors from Rag2c/c p53−/− and Atm−/− mice17 may arise not only as a result of Tcrd recombination (as shown previously in ATM-deficient mice16) but also from Tcra recombination. It has previously been proposed that the aberrant interlocus Tcra/d-Igh rearrangements found in Atm−/− T cells arise from the persistence of unrepaired breaks converging at different stages of thymocyte differentiation16. However, in contrast to our analyses, these studies did not examine breaks on Tcra/d and Igh in individual developing T cells, but rather they analyzed translocations in Atm−/− mature T cells and Atm−/− derived thymic lymphomas, respectively. Here we show that breaks on the two loci occur in the same cell at the same stage of development, which provides a direct mechanism for the generation of the characteristic inter-locus Tcra/d-Igh translocations that are found in Rag2c/c p53−/− and Atm−/−17,47,48 T-lymphomas.
Our studies reveal that there is similarity between the C-terminus of RAG2 and ATM in temporally harnessing RAG activity to ensure that cleavage occurs on only one locus at a time in recombining T cells. Intriguingly we find that RAG mediated association of recombining loci in localized ‘recombination centers’ is linked to feedback control of RAG cleavage: the introduction of a break on one locus is coupled with repositioning of the partner locus to PCH and inhibition of bi-locus cleavage and looping out from the territory (see Model in Supplementary Fig. S3). Targeting of RAG to the correct allele (Tcra rather than Igh) is likely to be influenced by the higher level of transcription on Tcra. Interestingly, although cleavage of Tcra is reduced in paired Tcra/d-Igh alleles in Rag2c/c p53−/− DP cells (perhaps due to a decrease in efficiency of cleavage in the absence of the C-terminus of RAG2), there is a significant increase in cleavage of Igh, which further underlines the role of regulation in trans between the two loci. Although we recently showed that pairing and higher-order looping of Tcra homologous alleles is linked with regulation of their recombination40, cross-talk and regulation of heterologous loci in trans has not previously been shown to be linked to the formation of higher-order looping. Indeed higher-order loop formation was previously shown to be involved in stochastic interactions between different loci on separate chromosomes41,49 while here we have found that interactions between Tcra/d and Igh are mediated by the presence of RAG.
Together our data suggest that regulation of mono-locus cleavage relies on changes in nuclear organization that are associated with a reduction in accessibility. Regulation of mono-locus cleavage is thus akin to regulation of mono-allelic cleavage. Indeed, our previous studies showed that ATM recruited to the site of a break on one allele acts in trans to reposition the second homologous allele to PCH and to prevent the introduction of further breaks on the partner homologue40,50. We propose that homologous and heterologous antigen receptor alleles come together in localized recombination centers for coordinated regulation of RAG cleavage. Here we have now identified an auto-regulatory role for RAG2 in restricting cleavage in an analogous manner to ATM. These data explain the mechanisms underlying the origins of the inter-locus Igh-Tcra/d in Atm−/− and Rag2c/c p53−/− lymphocytes prior to lymphomagenesis. Importantly, they identify an unappreciated role for the C terminal of RAG2 in regulating chromosome dynamics and accessibility of target loci to restrict ongoing cleavage after the introduction of a break on one locus. Thus feedback control of RAG activity relies on signals transmitted via the RAG complex itself.
Regulation of V(D)J recombination occurs at multiple levels to prevent the occurrence of chromosomal translocations or deletions that can result from errors in repair and/or mis-targeting of RAG1/2 to cryptic RSSs. However, beyond degradation of RAG2 protein8 and our more recent findings that expression of Rag1 is regulated across cell cycle51, there been have no studies that focus specifically on auto-regulation of RAG cleavage activity in individual cells. It is clearly critical to have such mechanisms in place to ensure that further breaks are not introduced in cis, or in trans on accessible target loci that undergo recombination at overlapping stages of development (Tcrg, Tcrb, Tcrd and Igh in DN T cells and Igk and Igl in pre-B/immature B cells). Furthermore, this same mechanism could also be important in preventing cleavage on actively transcribed off-target loci with cryptic RSSs that bind RAG.
Methods
Mice
The Atm−/−, Rag2c/c and Rag2c/c p53−/− mice were provided by Ludovic Deriano and David Roth17. RAG1-deficient mice were provided by Yanhong Ji, Grace Teng and David Schatz: RAG1-deficient DP cells (R1−/− β and control WT β) were derived from mice carrying a functionally rearranged Tcrb transgene (β) that allows T cell development to proceed to the DP stage in the absence of Tcrb rearrangement. The 53BP1−/− mice were provided by Davide Robbiani and Michel Nussenzweig52. Wild-type littermates were used as controls. Animal care was approved by NYU School of Medicine Animal Care and Use Committee of (protocol number 120315-01).
T cell flow cytometry sorting
Flow cytometry cell sorting was performed on a MoFlo or Reflection sorter. Antibodies were as follows: Thy1.2 PE-Cy7 (CD90.2, clone 53-2.1, eBioscience, 1:1000 dilution), TCRβ APC-eFluor780 (clone H57-597, eBioscience, 1:500 dilution), CD4 APC (L3T4, RM4-5, BD Biosciences, 1:500 dilution), CD8a FITC (clone 53-6.7, BD Biosciences, 1:500 dilution), CD25 PE (PC61, BD Biosciences, 1:500 dilution). The gating strategy was: Thy1.2+/TCRβint/CD4+/CD8+ for DP cells and Thy1.2+/TCRβlow/CD4−/CD8−/CD25+ for DN2/3 cells.
3-D DNA FISH and immuno-FISH
3D-DNA FISH and combined DNA FISH-immunofluorescence for γ-H2AX (immuno-FISH) were carried out on T cells adhered to poly-L lysine coated coverslips, as previously described50,53. Briefly, cells were fixed with 2% paraformaldehyde / PBS (pH 7-7.4) for 10 minutes at room temperature (RT) and permeabilized for 5 minutes with 0.4% Triton / PBS on ice. After 30 minutes of blocking in 2.5% BSA / 10% normal goat serum / 0.1% Tween-20 / PBS, cells were sequentially incubated with a primary antibody against phosphorylated serine-139 of H2AX (γ-H2AX; Millipore) and a secondary goat-anti-mouse antibody (Alexa Fluor 488 or 555; Invitrogen) for one hour at RT each. Cells were then post-fixed in 2% paraformaldehyde / PBS for 10 minutes at RT, incubated with 0.1 mg/ml RNaseA for 30 minutes at 37°C and permeabilized in 0.7% Triton-X-100 / 0.1M HCl for 10 minutes on ice. Cells were then denatured with 50% formamide / 2×SSC (pH 7-7.4) for 30 minutes at 80°C, and hybridized overnight with the probes at 37°C. The next day, cells were rinsed 3 times in 50% formamide / 2×SSC and 3 times in 2×SSC at 37°C for 5 minutes each. Finally slides were mounted in ProLong Gold (Invitrogen) containing 1.5 μg/ml DAPI.
Probes
BAC probes RP23-255N13 (3′α Tcra), RP23-304L21 (5′α Tcra), CT7-34H6 (3′ Igh), RP24-386J17 (5′ Igh), RP24-289O10 (Hmgb1), RP24-358H24 (Gapdh), RP24-342A2 (Lat), RP23-410N16 (Cd3g), RP24-277H9 (Ly6d), and RP23-137H17 (Satb1) were directly labeled by nick translation with ChromaTide Alexa Fluor 488 or 594-5-UTP (Molecular Probes) or Cy3- or Cy5-dUTP (Fisher). For one coverslip, 0.5 μg of nick-translation product was precipitated and resuspended in 10 μl of hybridization buffer (50% formamide / 20% dextran sulfate / 5× Denharts solution), denatured for 5 minutes at 95°C and pre-annealed for 45 minutes at 37°C before overnight hybridization with cells. XCyting Mouse Chromosomes 14 (Texas-red) and 12 (FITC) paints (Metasystems) were prepared separately following supplier's instructions. Paint and BAC probes were combined just prior to overnight hybridization.
Confocal microscopy and analysis
3-D images were acquired by confocal microscopy on a Leica SP5 AOBS system (Acousto-Optical Beam Splitter). Optical sections separated by 0.3 μm were collected and stacks were analyzed using Image J software. Alleles were defined as associated with γ-H2AX if the BAC signals and immunofluorescence foci were at least partially overlapped (at least one pixel of colocalization). Alleles were considered as located at pericentromeric heterochromatin (PCH) when BAC signals were adjacent or overlapping with a γ-satellite signal (no pixel in between the edges of the BAC and γ-satellite signals). Distances from the loci to their chromosome territories (higher-order looping) were measured from the centre of mass of the BAC signal to the closest edge of the chromosome paint. Distances between alleles or loci were measured between the center of mass of each BAC signal.
Statistical analyses
The statistical tests were applied to combined data sets from repeated experiments. Supplementary tables display individual experiments to show the low level of variation between the repeats. Statistical analyses were performed using a two-tail Fisher-exact test: P-values ≤ 5.00e-2 (a = 0.05) were taken to be significant (1.00e-2 ≤ P ≤ 5.00e-2 * significant; 1.00e-3 ≤ P ≤ 1.00e-2 ** very significant; P < 1.00e-3 *** highly significant).
Circularized chromosome conformation capture (4C-seq)
4C-seq and domainogram analyses were performed as previously described35,40,54. Fixation and cell lysis - 107 cells were resuspended in 5ml PBS-10% FBS and fixed in 5ml 4% formaldehyde-10%FBS for 10min at RT (tumbling). 1.425ml 1M glycine was added to quench cross-linking and tubes were put on ice for 2min. After 8min centrifugation at 1300RPM at 4°C, pellets were resuspended in 1ml cold “lysis buffer” (50mM Tris pH7.5, 150mM NaCl, 5mM EDTA, 0.5% NP-40, 1% Triton X-100, 1 tablet mini complete) and incubated for 20min on ice. After 5min centrifugation at 1800RPM at 4°C, pellets were resuspended in 360μl sterile water (or stored at −80°C). HindIII digestion - Nuclei were incubated for 1h at 37°C with 60μl 10× restriction buffer B and 15 μl 10% SDS (shaking). 150 μl 10% Triton X-100 were added and nuclei were incubated for 1h at 37°C (shaking). 5 μl of the sample were taken as the “undigested control”. Nuclei were incubated overnight at 37°C with 400U of HindIII restriction enzyme. Fresh enzyme was added for 6 more hours during the day. 5 μl “undigested” and “digested” controls were incubated in 90 μl Tris pH7.5 and 5 μl 10mg/ml Proteinase K for 2h at 65°C, DNA was extracted with 100 μl phenol-chloroform and the water phase was loaded on a 0.6% agarose gel. The HindIII enzyme was inactivated by adding 80 μl 10% SDS and incubating at 37°C for 30 min. Ligation – Samples were transferred into 15ml falcons and incubated with 4860 μl sterile water, 700 μl 10× ligation buffer and 750 μl 10% Triton X-100 for 1h at 37°C (shaking). 50U ligase were added and incubated overnight at 16°C. 100 μl of the sample were taken as the “ligated control”, tested as above (5 μl of 10mg/ml Proteinase K for 2h at 65°C and phenol extraction), and compared to the “digested control” on a 0.6% agarose gel.De-crosslinking and precipitation – De-crosslinking was performed by adding 30 μl of 10mg/ml Proteinase K for 4h at 65°C. After 45min incubation in 30 μl RNase A (10mg/ml) at 37°C, DNA was phenol extracted and ethanol precipitated and resuspended in 150 μl 10mM Tris pH 7.5. DpnII digestion - HindIII-ligated 3C template was digested overnight at 37°C with DpnII (50 μl 10× restriction buffer, 50U DpnII, 300μl sterile water). DpnII was inactivated at 65°C for 30min. Ligation and precipitation – DNA was ligated overnight at 16°C in 12ml sterile water, 1.4ml ligation buffer, and 100U ligase. Ligation products were phenol extracted and ethanol precipitated using glycogen as a carrier (20 mg/ml), and resuspended in 75 μl 10 mM Tris pH7.5. Samples were purified using Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare). PCR reaction and sequencing - Specific primers for the Tcra Ea enhancer were HindIII AGACAGACCCTGCGAAGCTT and DpnII TAAGACTGGACCCACAG. The Illumina-specified adapters for Illumina GAIIx sequencing were included at the 5′ end of each primer. PCR reactions were performed using the Expand Long Template PCR system (Roche) and PCR conditions were as following: 94°C for 2 min; 30 cycles of 94°C for 1min, 55°C for 1min and 68°C for 3min; followed by a final step of 68°C for 7min. The 4C library was sequenced on an Illumina GAIIx single-read 72-cycle run. 4C-seq analysis - the 54bp single-end reads were aligned to a library of 40bp flanking regions of HindIII restriction sites, on the mouse genome sequence (build mm9). The alignment was performed using Maq software (http://maq.sourceforge.net/) with a quality threshold of 150. We kept the distinction between the upstream and downstream flanking region of each HindIII site. We segmented the genome according to the HindIII restriction sites and removed those segments consisting of two consecutive HindIII restriction sites that did not contain a DpnII restriction site. Domainogram analysis –We followed the analytical steps described previously38, using the genomic regions enclosed between two contiguous HindIII sites. We then used windows of increasing size extending up to 30 HindIII sites. We computed and analyzed scores in each window size as follows: first we scored each pair of contiguous HindIII sites (defined as Q(x) = (rank(x)−0.5)/N, where Q is a quantile score of HindIII sites (x) and N is the total number of these sites). The Qi values are calculated in a global manner for the whole genome, with the bait chromosome analyzed separately. We transformed the combination of scores within each window to a form that was amenable to applying Fisher's approach for combining independent tests of significance. The range of colors in the domainograms represents the intensity of these scores. See Supplementary Table S4 for details.
ChIP-seq
ChIP-seq preparation was carried out as previously described55 and ChIP-seq analysis was performed using the Qeseq algorithm56 (details were provided in Chaumeil et al40). Cell fixation and lysis - 1-5×106 cells were fixed in 1% formaldehyde for 10 min at RT and lysed in 15mM Tris pH 7.5, 60mM KCl, 15mM NaCl, 15mM MgCl2, 1mM CaCl2, 250mM Sucrose, and 0.3% NP-40 for 10min at 4°C. Nuclei were isolated by centrifugation and washed once in “digest buffer” (10mM NaCl, 10mM Tris-HCl pH 7.5, 3mM MgCl2, 1mM CaCl2, 0.1 mM PMSF). Chromatin preparation - Mononucleosomal particles were generated using Micrococcal nuclease (USB) in “digest buffer” (reaction stopped with 20mM EDTA). Nuclei were then lysed in “nucleus lysis” buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA (pH 8.0) and 1% SDS) and sonicated (2.5 min total; Bioruptor (Diagenode)). Chromatin was pre-cleared by addition of nine volumes of “IP dilution” buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA (pH 8.0), 16.7 mM Tris-HCl pH 8.0 and 167 mM NaCl) and magnetic Dynal beads. 1% of chromatin was saved as “input”. Chromatin immunoprecipitation - Antibodies (H3K4me3, Active motif; H3K9ac; Abcam) and beads were incubated together for 4 hours in “IP dilution” buffer, before overnight incubation with the chromatin at 4°C, while rotating. The complexes bound on the beads were then washed in buffers with increasing salt concentration (“wash A”: 20 mM Tris-HCl pH 8, 150 mM NaCl, 2 mM EDTA, 1% w/v Triton, 0,1% w/v SDS; “wash B”: 20 mM Tris-HCl pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% w/v Triton, 0.1% w/v SDS; “wash C”: 10 mM Tris-HCl pH8.0, 250 mM LiCl, 1 mM EDTA, 1% w/v Nonidet P-40, 1% w/v deoxycholic acid) and twice with TE. The bound chromatin was cleaned using Proteinase K, at 65°C overnight and DNA was phenol extracted and ethanol precipitated. ChIP-seq libraries and sequencing - ChIP-seq libraries were generated using standard Illumina kit protocol. Truseq adapters were added and the libraries were PCR-amplified. Size selection was performed using E-gel electrophoresis system by Invitrogen. Cluster amplification and 36-nucleotide single-end sequencing on an Illumina Genome Analyzer II were performed following manufacturer's instructions. ChIP-seq analysis - Read were aligned with Bowtie 0.12.7 software to the reference mouse genome (mouse assembly NCBI mm9), using the following command line option-best-all –m1 –n2 (reads that align uniquely in the best alignment stratum, allowing up to two mismatches).See Supplementary Table S5 for details.
Western blotting
Cells were incubated in lysing buffer (0.2% sodium dodecyl sulfate, 100mM Tris-HCL) at 16,000 cells per μl for 5 min at 95°C and treated with Bensonaze nuclease (0.05U/μl) for removal of nucleic acids for 5 min at room temperature. Protein extracts were denatured and reduced in 1× Nupage ® sample LDS and 1× Nupage ® sample reducing agent (Life Technologies) for 5 min at 95°C before the equivalent of 2 × 105 cells per lane was loaded. Membranes were blocked in phosphate-buffered saline (PBS) with 5% milk and 0.1% Tween and incubated with the following primary antibodies in 5% milk and 0.1% Tween: ATM (MAT3-4G10/8; 1/4000, Abcam) and γ-Tubulin (clone GTU-88; 1/5000, Sigma), which was used as a loading control. Blots were developed with enhanced chemiluminescence (Pierce).
Supplementary Material
Acknowledgments
The authors thank D. Schatz, Y. Ji and G. Teng for providing the RAG1-deficient mice, and M. Nussenzweig and D. Robbiani for the 53BP1-deficient mice. We would like to thank J. Allinne, S. Hewitt, C. Proudhon, R. Raviram, P. Rocha, M. Sellars, and T. Trimarchi for their help with 4C-seq experiments and analyses; E. Nora for his help with Image J; and members of the Skok lab for thoughtful discussions and comments on the study. This work is supported by the National Institute of Health: RO1GM086852 and R56NIAIDAI099111 (J.A.S); CA-16359 (Y.K.); RO1CA133379, RO1CA105129, RO1CA149655, RO1CA173636 and RO1GM088847 (I.A.); and CA104588 (D.B.R.). J.A.S is an LLS scholar. J.C. is an Irvington Institute Fellow of the Cancer Research Institute. M.M. was supported by an NSF IGERT 0333389. P.N. is supported by the Lady Tata Memorial Trust for Leukemia. I.A. is also supported by the Leukemia & Lymphoma Society (TRP grant), the Chemotherapy Foundation, the V Foundation of Cancer Research, and the William Lawrence Blanche Hughes Foundation, and is an early Career Scientist of the Howard Hughes Medical Institute. L.D. is supported by the Institut Pasteur, the CNRS, the Fondation pour la Recherche Medicale, the Ville de Paris, and by the European Research Council (ERC starting grant agreement #310917).
Footnotes
Author Contribution: JC and JS conceived the study and wrote the manuscript. JC performed most of the experiments. MM analyzed the 4C-seq and ChIP-seq data. PN performed the ChIP-seq experiments. LD analyzed the damage experiments, performed the ATM western blot, provided Atm−/−, Rag2c/c and Rag2c/c p53−/− mice and revised the manuscript. DBR, IA and YK provided technical and conceptual support. All the authors read and approved the manuscript.
Competing Interests Statement: The authors declare that they have no competing financial interests.
References
- 1.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 2.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 4.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 7.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 11.Puebla-Osorio N, Zhu C. DNA damage and repair during lymphoid development: antigen receptor diversity, genomic integrity and lymphomagenesis. Immunol Res. 2008;41:103–122. doi: 10.1007/s12026-008-8015-3. [DOI] [PubMed] [Google Scholar]
- 12.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciofani M, Zuniga-Pflucker JC. Determining gammadelta versus alphass T cell development. Nat Rev Immunol. 2010;10:657–663. doi: 10.1038/nri2820. [DOI] [PubMed] [Google Scholar]
- 14.Livak F, Tourigny M, Schatz DG, Petrie HT. Characterization of TCR gene rearrangements during adult murine T cell development. J Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- 15.Kurosawa Y, et al. Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 1981;290:565–570. doi: 10.1038/290565a0. [DOI] [PubMed] [Google Scholar]
- 16.Zha S, et al. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. J Exp Med. 2010;207:1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deriano L, et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang HE, et al. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 19.Jones JM, Simkus C. The roles of the RAG1 and RAG2 “non-core” regions in V(D)J recombination and lymphocyte development. Arch Immunol Ther Exp (Warsz) 2009;57:105–116. doi: 10.1007/s00005-009-0011-3. [DOI] [PubMed] [Google Scholar]
- 20.Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Molecular and cellular biology. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Molecular cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 22.Yarnell Schultz H, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Molecular cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 23.Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic acids research. 2004;32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 25.Curry JD, Schlissel MS. RAG2's non-core domain contributes to the ordered regulation of V(D)J recombination. Nucleic acids research. 2008;36:5750–5762. doi: 10.1093/nar/gkn553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bredemeyer AL, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 27.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 28.Chen HT, et al. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callen E, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 31.Callen E, Nussenzweig MC, Nussenzweig A. Breaking down cell cycle checkpoints and DNA repair during antigen receptor gene assembly. Oncogene. 2007;26:7759–7764. doi: 10.1038/sj.onc.1210873. [DOI] [PubMed] [Google Scholar]
- 32.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborne CS, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocha PP, et al. Close Proximity to Igh Is a Contributing Factor to AID-Mediated Translocations. Molecular cell. 2012;47:873–885. doi: 10.1016/j.molcel.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bantignies F, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 39.Ji Y, et al. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaumeil J, et al. Higher-order looping and nuclear organization of Tcra facilitate targeted rag cleavage and regulated rearrangement in recombination centers. Cell reports. 2013;3:359–370. doi: 10.1016/j.celrep.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nature immunology. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 45.Splinter E, et al. The inactive × chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liyanage M, et al. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]
- 48.Callen E, et al. Chimeric IgH-TCRalpha/delta translocations in T lymphocytes mediated by RAG. Cell Cycle. 2009;8:2408–2412. doi: 10.4161/cc.8.15.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheutin T, Cavalli G. Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion. PLoS genetics. 2012;8:e1002465. doi: 10.1371/journal.pgen.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hewitt SL, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nature immunology. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson K, et al. IL-7 functionally segregates the pro-B cell stage by regulating transcription of recombination mediators across cell cycle. J Immunol. 2012;188:6084–6092. doi: 10.4049/jimmunol.1200368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Molecular and cellular biology. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaumeil J, Micsinai M, Skok JA. Combined immunofluorescence and DNA FISH on 3D-preserved interphase nuclei to study changes in 3D nuclear organization. Journal of visualized experiments : JoVE. 2013:e50087. doi: 10.3791/50087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature genetics. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 55.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asp P, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E149–158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.