FIG 2 .
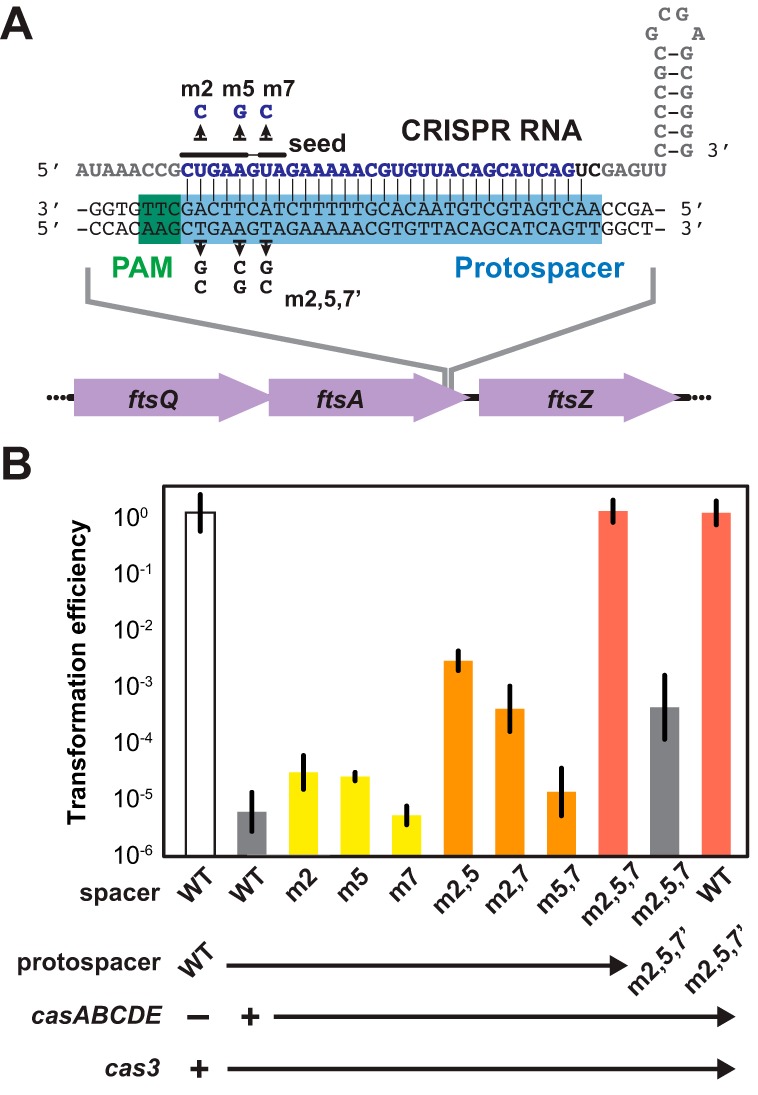
Potent and sequence-specific removal through genome targeting with CRISPR-Cas systems. (A) Design of a CRISPR RNA targeting the ftsA gene in E. coli K-12. The 32-nt spacer sequence is in blue, and the repeat sequence is in gray. The last two nucleotides of the spacer (in black) are fixed to introduce restriction sites used for cloning additional repeat-spacer pairs. Flanking the protospacer (highlighted in blue) is the protospacer-adjacent motif (PAM) (highlighted in green) required for DNA targeting. Point mutations within the established seed region of the spacer (9) and the protospacer tested in panel B are shown. (B) Transformation efficiencies of α-ftsA plasmids containing different mutations in the seed region of the spacer. Single, double, and triple mutations of the spacer sequence are shown in yellow, orange, and red, respectively. The transformations were carried out in BW25113-T7 (wild type [WT]) or BW25113-T7m257′ (m2,5,7′), each harboring two plasmids: pCas3 (+ cas3) and either pCasA-E (+ casABCDE) or pCasA-E′ (− casABCDE). Figure S1 in the supplemental material illustrates the general transformation procedure. Transformation efficiency was calculated as the number of transformants for each tested plasmid divided by the number of transformants for the original pCRISPR plasmid for the same culture. Values represent the geometric means and SEM of data from three independent experiments.
