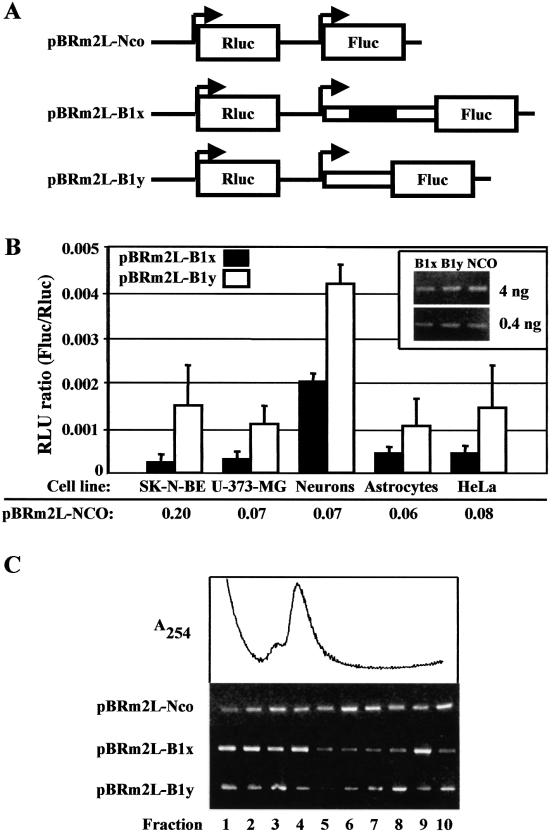
Figure 3.
Translation driven by the BACE-1 transcript leaders. (A) Scheme of bi-monocistronic constructs. The plasmid pBRm2L-Nco drives expression of the two reporter genes, Rluc and Fluc, under the control of separate T7 promoters and in an optimal context for translation initiation. The long and short BACE-1 transcript leaders were cloned in front of the Fluc ORF to generate pBRm2L-B1x and pBRm2L-B1y, respectively. (B) Transient transfection of pBRm2L-B1x or pBRm2L-B1y in SK-N-BE, U-373-MG, neurons, astrocytes and HeLa cells. To account for differences in transient transfection efficiencies, the activity (relative luminescence units, RLU) of Fluc was normalized to that of Rluc (Fluc/Rluc). The corresponding RLU ratio values for the empty vector pBRm2L-Nco are also shown for comparison. All the results are the mean of at least three independent experiments. Measurements of luciferase activity were performed in triplicate as described in Materials and Methods. The inset shows an RT–PCR experiment (with two different total RNA loadings) to evaluate the relative amount of Fluc transcripts in transfected HeLa cells. The efficiency of transfection was monitored by Rluc activity and proved to be comparable in all samples. (C) Cytoplasmic distribution of Fluc mRNA by sucrose gradients. Cytoplasmic extracts of transfected HeLa cells were lysed and resolved on 7–50% w/v sucrose gradients. Fractions were analysed by RT–PCR to reveal the localization of the transcript deriving from the transfected DNA (pBRm2L-Nco, pBRm2L-B1x or pBRm2L-B1y). The trace shows the continuous absorbance profile monitored during the collection of the fractions. Fractions 1–3 contain mainly mRNPs and the ribosomal subunits; the monosome region peaks at fraction 4; fractions 5–10 contain the polyribosomes.