The authors performed metabolomics analysis on a time series of guard cell samples treated with abscisic acid (ABA) to follow the dynamics of 85 metabolites potentially involved in ABA signaling. ABA rapidly altered the profiles of numerous metabolites, including other hormones and calcium-mobilizing agents. ABA responses were greater in the wild type than in a G-protein mutant with ABA-hyposensitive stomata.
Abstract
Individual metabolites have been implicated in abscisic acid (ABA) signaling in guard cells, but a metabolite profile of this specialized cell type is lacking. We used liquid chromatography–multiple reaction monitoring mass spectrometry for targeted analysis of 85 signaling-related metabolites in Arabidopsis thaliana guard cell protoplasts over a time course of ABA treatment. The analysis utilized ∼350 million guard cell protoplasts from ∼30,000 plants of the Arabidopsis Columbia accession (Col) wild type and the heterotrimeric G-protein α subunit mutant, gpa1, which has ABA-hyposensitive stomata. These metabolomes revealed coordinated regulation of signaling metabolites in unrelated biochemical pathways. Metabolites clustered into different temporal modules in Col versus gpa1, with fewer metabolites showing ABA-altered profiles in gpa1. Ca2+-mobilizing agents sphingosine-1-phosphate and cyclic adenosine diphosphate ribose exhibited weaker ABA-stimulated increases in gpa1. Hormone metabolites were responsive to ABA, with generally greater responsiveness in Col than in gpa1. Most hormones also showed different ABA responses in guard cell versus mesophyll cell metabolomes. These findings suggest that ABA functions upstream to regulate other hormones, and are also consistent with G proteins modulating multiple hormonal signaling pathways. In particular, indole-3-acetic acid levels declined after ABA treatment in Col but not gpa1 guard cells. Consistent with this observation, the auxin antagonist α-(phenyl ethyl-2-one)-indole-3-acetic acid enhanced ABA-regulated stomatal movement and restored partial ABA sensitivity to gpa1.
INTRODUCTION
Owing to advances in mass spectrometry, metabolite profiling for metabolomic analyses has advanced significantly over the last decade. In nontargeted liquid chromatography–tandem mass spectrometry (LC-MS/MS) approaches, metabolites are characterized by the specific MS/MS spectra that they generate. In some cases, a spectrum from the sample of interest will match that of a known metabolite (Oliver et al., 2011), thereby allowing explicit identification. In many other cases, it will not be possible to associate spectra with a named metabolite. Nontargeted metabolite profiling thus provides a high-throughput large-scale overview of the metabolome (Last et al., 2007; Böttcher et al., 2008; Allwood and Goodacre, 2010; Oliver et al., 2011). An alternative approach is targeted metabolomics, wherein a unique LC-MS/MS profile for each metabolite of interest is first generated using authentic chemical standards. These metabolites are then identified in the sample of interest and quantified by reference to the known standards. Targeted metabolomics enables high sensitivity, high selectivity, and large dynamic range detection of metabolites as well as absolute quantification (Shulaev et al., 2008; Allwood and Goodacre, 2010). One limitation of both nontargeted and targeted metabolomics, in contrast with proteomics and transcriptomics, is the current machine dependency of the data, such that it is difficult to generate universally applicable spectral libraries for LC-MS/MS data. Nevertheless, and despite the fact that only a minority of the estimated ∼200,000 plant metabolites (Goodacre et al., 2004) have been identified thus far, many of the known plant secondary metabolites have nutritional or medicinal benefits (Hirai et al., 2004; DellaPenna and Last, 2008).
Increasing attention on stress-regulated metabolomes has advanced our understanding of plant metabolic reprogramming and acclimation under various environmental conditions including those relevant to abscisic acid (ABA) such as drought/dehydration, salinity, and temperature stress. By detaching rosettes of wild-type Arabidopsis thaliana plants versus those of the ABA biosynthesis mutant nc3-2 and allowing them to dehydrate while sampling over a 15-h time course, Urano et al. (2009) revealed ABA-dependent and independent responses of the nontargeted metabolome, including ∼100 identified metabolites. Consistent with the conclusion that ABA plays important roles in various environmental stress responses but that there are also intrinsic stress-specific pathways that are ABA independent, metabolite profiling of leaves from 4-week-old Arabidopsis plants watered with 150 mM NaCl or sprayed with 25 µM ABA for 2 h to 3 d revealed starch depletion and strong elevation of maltose by both ABA and high salt stress, but differential stress regulation of other soluble carbohydrates (Kempa et al., 2008). Gas chromatography–mass spectrometry quantification of 52 polar compounds in Arabidopsis leaf samples revealed that the metabolome resulting from a combined drought and heat stress shared more similarity with a drought-stressed metabolome than with a heat-stressed metabolome (Rizhsky et al., 2004). Studies of ABA- and dehydration-responsive metabolomes also have been conducted in rice (Oryza sativa) suspension culture cells (Rao et al., 2010), in leaves from potato (Solanum tuberosum) cultivars subjected to drought in the field (Evers et al., 2010), and in leaves from Sporobolus grass species differing in desiccation (Oliver et al., 2011).
Although there is increasing application of metabolomics in cell culture and whole plant or whole organ responses to abiotic stresses, only a few single cell types of vascular plants have been subjected to detailed metabolite investigation (Rubakhin et al., 2013). Schilmiller et al. (2010) employed gas chromatography–mass spectrometry and liquid chromatography/time-of-flight mass spectrometry on isolated trichomes to profile volatile and nonvolatile metabolite phenotypes of tomato (Solanum esculentum) chromosomal introgression lines harboring chromosomal regions of the wild relative, Solanum penellili, and found differences in accumulation of terpenes, acyl sugars, and acyl sugar metabolites. Ebert et al. (2010) developed a gas chromatography/time-of-flight mass spectrometry method for identification of ∼100 metabolites from pools of 200 microcapillary-sampled epidermal cells and found that 38 metabolites displayed different pool sizes in trichomes, pavement cells, and the basal cells surrounding trichomes. Because of high standard deviation and detection limits, the other ∼62 metabolites could not be quantified, and the authors proposed that analysis of much larger numbers of cells would be beneficial for improving detection and reliability (Ebert et al., 2010). Alternatively, in situ sampling methods hold promise for querying of single cell metabolomes. For example, using a nanoelectrospray tip to sample individual geranium (Pelargonium zonale) petal, leaf, and stem cells, ∼20 compounds, primarily alcohols and terpenoids, were identified by electrospray ionization–tandem mass spectrometry (ESI-MS/MS) (Lorenzo Tejedor et al., 2012); however, it is difficult to identify more metabolites and achieve quantitative analysis. In another approach, laser ablation ESI-MS was applied to onion (Allium cepa) epidermal cells and sour orange (Citrus aurantium) oil glands and ∼10 metabolites were identified, primarily sugars and flavonoids (Shrestha et al., 2011). Because the reported sampling diameter and depth were both 30 µm, this approach would not be applicable for single-cell sampling of small cells such as guard cells.
The above investigations indicate the challenge of profiling the metabolomes of single cell types using advanced mass spectrometric techniques. Guard cells are a single cell type found in the epidermis of land plants, where they regulate stomatal apertures through which carbon dioxide uptake for photosynthesis and loss of water vapor and oxygen release occur. Guard cells have evolved exquisite mechanisms to sense and respond to endogenous and environmental stimuli such as plant hormones, including ABA, as well as to plant water status, light, carbon dioxide, temperature, and humidity. Guard cells have become a premier model system for studies of cellular signaling (Assmann, 2010) in large part because the effects of a stimulus on guard cell ion channels, cytosolic ion concentrations, and stomatal apertures can be assayed in real time, and it is also feasible, albeit challenging, to isolate large populations of highly pure guard cells for transcriptomic (Leonhardt et al., 2004; Pandey et al., 2010; Wang et al., 2011) and proteomic (Zhao et al., 2008, 2010; Zhu et al., 2009, 2010) analyses. Dozens of individual cellular compounds and processes, including osmolytes, lipid metabolites, cyclic nucleotides, elevation of cytosolic free calcium concentration ([Ca2+]cyt), and production of reactive oxygen species (ROS) and nitric oxide (NO), have been implicated in ABA-responsive guard cell signaling pathways (MacRobbie, 1998; Lamattina et al., 2003; Hetherington and Brownlee, 2004; Li et al., 2006; Wang and Song, 2008; Kim et al., 2010; Roelfsema and Hedrich, 2010). To date, however, little is known about the guard cell metabolome; rather, each identified metabolite component of the signaling network has been implicated by largely independent studies utilizing biochemical assays, pharmacological interventions, or genetic manipulations. Separate studies on individual metabolites cannot reveal covariance/correlation matrices of metabolites that may be indicative of metabolite crosstalk. Accordingly, application of metabolomics approaches to access the simultaneous analysis of many metabolites over time is required to achieve insights into the guard cell metabolome.
Steady state metabolite analysis achieved by sampling at beginning and end points can provide at best limited temporal information, particularly because linearity in metabolite concentration changes over time cannot be assumed (Stitt et al., 2010). Until now, very few time-series experiments on plant metabolomes have been conducted (Kim et al., 2007; Hoefgen and Nikiforova, 2008; Kempa et al., 2008; Rao et al., 2010), and these studies often have profiled samples collected over relatively large time intervals (hours) and after prolonged stress (up to several days). By contrast, guard cells can respond to stimuli within seconds (Assmann and Grantz, 1990) and exhibit reversible responses that are a key aspect of dynamic stomatal regulation (Israelsson et al., 2006; Shimazaki et al., 2007; Acharya and Assmann, 2009). Therefore, short-term time series experiments are needed to unravel the signaling metabolome of this specific cell type.
Here we performed metabolomic analysis on a time series of guard cell samples in order to follow the dynamics of a number of metabolites potentially involved in ABA regulation of guard cell signaling. We utilized the technique of LC-MS/MS with multiple reaction monitoring (MRM), in which metabolites are uniquely identified by matching their precursor/product (parent/daughter) ion pairs to those of authentic chemical standards. We chose to employ a targeted metabolomics approach because of our interest in quantitatively investigating the coregulation of a number of metabolites previously implicated in guard cell ABA response, and because this approach enabled us to compare these metabolites in wild-type Arabidopsis ecotype Columbia (Col) plants versus heterotrimeric GTP binding protein (G protein) α subunit mutant (gpa1) plants. Heterotrimeric G proteins are composed of a Gα, Gβ, Gγ trimer that dissociates into the signal transducing elements Gα and the Gβγ dimer upon activation, and are ubiquitous secondary messengers in eukaryotes (Perfus-Barbeoch et al., 2004). GPA1 is the sole canonical Gα subunit of Arabidopsis. Arabidopsis gpa1 mutants have been studied widely regarding physiological aspects, wherein gpa1 plants have been found to be hyposensitive in many aspects of guard cell ABA response, including ABA inhibition of stomatal opening, ABA inhibition of inward K+ channels, and ABA activation of calcium-permeable channels and production of ROS, but exhibit largely wild-type behavior for ABA promotion of stomatal closure (Wang et al., 2001; Fan et al., 2008; Zhang et al., 2011). gpa1 mutants also have reduced stomatal density in rosette leaves, which contributes to reduced whole leaf stomatal conductance and increased transpiration efficiency (Nilson and Assmann, 2010). We were interested in assessing GPA1 participation in ABA regulation of the guard cell metabolome.
Our targeted metabolomic analysis was performed on a total of ∼350 million guard cell protoplasts harvested from ∼30,000 Arabidopsis Col and gpa1 plants and subjected to a time series consisting of 0, 2, 10, 30, and 60 min of 50 µM ABA or ethanol (solvent control) treatment. Multiple replicates were assessed, with each replicate containing 3 to 6 million protoplasts for a total of 72 samples analyzed. We found that the guard cell ABA-regulated metabolome contains both known and heretofore unknown ABA signaling elements, and that many of these are coregulated by GPA1. Assessment in mesophyll cell protoplasts of a hormone-related subset of the metabolites highlighted the unique nature of the guard cell metabolome response to ABA.
RESULTS
Targeted Analysis of the Guard Cell Metabolome
A total of 85 metabolites, including hormones, lipid metabolites, flavonoids, polyphenols, amino acids, and carbohydrates, were quantified in Arabidopsis guard cells. We particularly focused on metabolites known or implicated in guard cell ABA response (Li et al., 2006). Plant hormones are obvious targets of interest and although ABA regulation of stomatal aperture has received predominant attention, other hormones also can affect stomatal apertures. Moreover, in other systems, hormones such as gibberellins, cytokinins, and auxin can alter the effectiveness of ABA (Davies, 2011). In guard cells, lipid metabolites such as inositol phosphates (Lee et al., 1996; Hunt et al., 2003; Lemtiri-Chlieh et al., 2003) and phosphorylated sphingolipids (Ng et al., 2001; Coursol et al., 2003; Coursol et al., 2005) are implicated as second messengers in ABA signaling. These lipid metabolites, as well as the cyclic nucleotide, cyclic adenosine diphosphate Rib (cADPR) (Leckie et al., 1998) elevate [Ca2+]cyt in guard cells in response to ABA, and elevated [Ca2+]cyt in turn inhibits K+ influx channels and activates anion efflux channels, contributing to inhibition of stomatal opening and promotion of stomatal closure, respectively. The cyclic nucleotide cGMP also participates in [Ca2+]cyt regulation in response to NO and ROS in guard cells (Garcia-Mata et al., 2003; Dubovskaya et al., 2011; Joudoi et al., 2013). ABA-induced production of ROS (Kwak et al., 2003) is important in the activation of guard cell ion transporters including Ca2+ uptake channels and anion release channels, and gpa1 mutants are deficient in ABA-induced ROS production (Zhang et al., 2011). We therefore also focused on characterizing metabolite regulators of cellular redox status, including antioxidants such as ascorbate and GSH (Chen and Gallie, 2004; Foyer and Noctor, 2011), as well as phenolic acids and flavonoids, which play important roles as reducing agents and free radical scavengers (Hichri et al., 2011; Kusano et al., 2011).
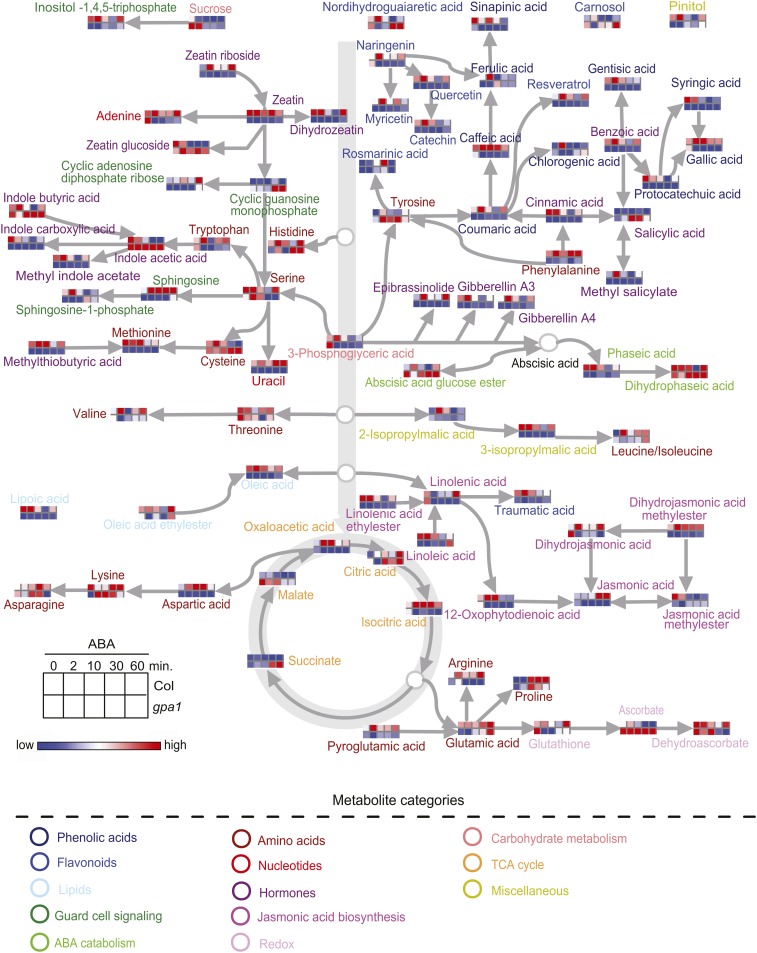
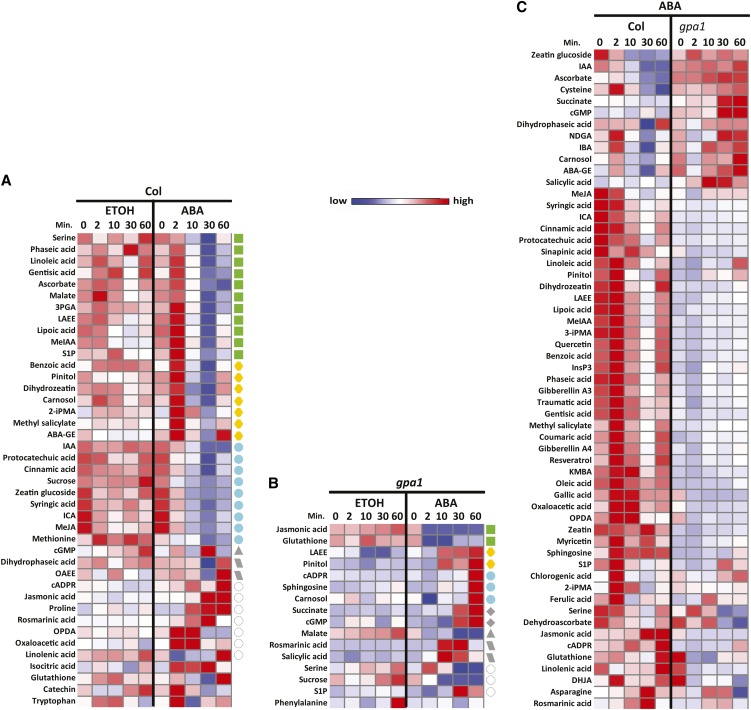
To provide an overview of the 85 metabolites (see Supplemental Data Set 1 online) and their inter-relationships, we mapped the 85 metabolites onto general biochemical pathways (Figure 1) as illustrated in the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) and the Plant Metabolic Network (http://www.plantcyc.org/). Inter-genotype (Col versus gpa1) comparisons over time of ABA treatment are presented as heatmaps in Figure 1 in order to provide a readily accessible and synthetic overview of the data. Statistically significant differences are identified and discussed in subsequent figures.
Figure 1.
An Overview of the Metabolic Pathways in which the Targeted Metabolites Are Located, and Their Responsiveness to ABA in Col and gpa1 Guard Cells.
The abundance of each metabolite under ABA treatment is represented by a heatmap. Solvent control (ethanol) treatments had essentially no effect (see text) and are not depicted with heatmaps. Metabolite names are color coded according to the categories to which they belong. It is important to note that arrows in this overview do not indicate direct interconversions between two adjacent metabolites but simply indicate overall upstream/downstream relationships in metabolic pathways.
Principal Component Analysis Indicates ABA and Genotypic Effects on the Guard Cell Metabolome
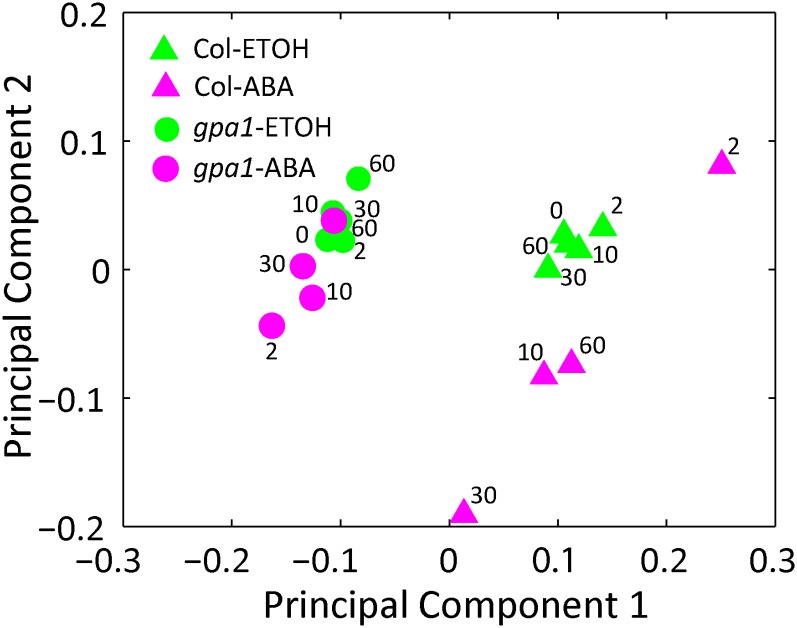
Principal component analysis (PCA) was employed to provide an overview of sample distribution. The score plot of the PCA of our guard cell metabolite data averaged over the replicates is shown in Figure 2, and PCA loadings are provided in Supplemental Data Set 2 online. Guard cell protoplast samples from Col treated with the solvent control, ethanol, grouped together with each other and with the Col zero time point, indicating that ethanol did not cause dramatic changes in metabolite abundance in Col. The same was found for gpa1 guard cell protoplasts: ethanol-treated samples cluster together with the gpa1 zero time point. Col data at 10 and 60 min after ABA treatment are separated from the Col controls, and Col data at 2 and 30 min of ABA treatment are particularly distant, indicating that the abundance of some metabolites changed dramatically after treatment with ABA at 2 min and 30 min (confirmed by subsequent analysis of variance [ANOVA], see below). The ABA-treated gpa1 samples are somewhat separate from their respective ethanol controls except for the final 60-min time point, suggesting a more moderate and transient metabolome response than seen for Col. In PCA (Figure 2), Col-ABA samples were more divergent from Col-ethanol samples than were gpa1-ABA samples from gpa1-ethanol samples, suggesting strong regulation of the ABA-responsive metabolome by the heterotrimeric G protein, which is consistent with previously documented ABA hyposensitivity of gpa1 guard cells in other aspects of stomatal function (Wang et al., 2001; Fan et al., 2008; Pandey et al., 2010; Wang et al., 2011; Zhang et al., 2011).
Figure 2.
PCA Scores Are Affected by Genotype and ABA Treatment.
The PCA score plot of the four genotype treatment conditions over all of the metabolites in guard cells is depicted. ETOH, ethanol.
[See online article for color version of this figure.]
ABA Causes Temporal Changes in Metabolite Abundance in Guard Cells
ANOVA was performed to assess whether the abundance of metabolites within a genotype changed significantly (P < 0.01) over time when subjected to either ethanol or ABA. Among the 85 metabolites quantified, only one metabolite, Leu/Ile, showed abundance changes in Col with respect to time after treatment with ethanol. Furthermore, only 2 of 85 metabolites in gpa1 (myricetin and linolenic acid ethylester) showed abundance changes with respect to time after treatment with ethanol. The scarcity of significant changes induced by the solvent control not only indicates that our ABA effects can be attributed to ABA rather than to the ABA solvent, ethanol, but also importantly suggest that our guard cell protoplasts were physiologically s\ and healthy during the 1-h treatment period. This conclusion is consistent with cell viability assessment using the vital stain fluorescein diacetate wherein >95% of guard cell protoplasts were found to be healthy and viable for up to 3 h (see Supplemental Figure 1 online). In addition, measurement of guard cell protoplast diameters over a 60-min time course of ABA or ethanol treatment demonstrated guard cell volume decrease in response to ABA, confirming ABA responsiveness (see Supplemental Figure 2 online).
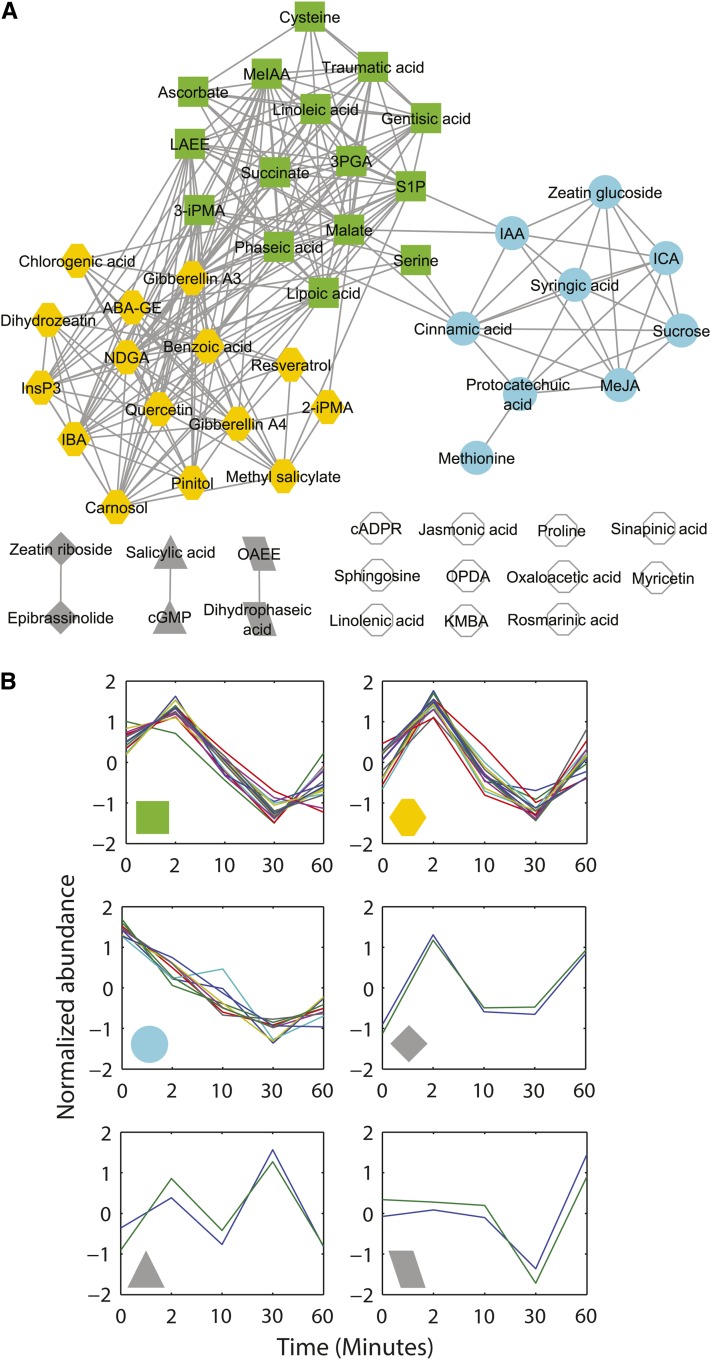
ANOVA indicated that 56 of 85 metabolites in Col showed significant changes over time when treated with ABA. We calculated the Pearson correlation coefficients (PCCs) of the variation over time of pairs of these 56 metabolites and connected two metabolites if their PCC was larger than 0.9. The resultant metabolite correlation network is shown in Figure 3A and demonstrates that 45 metabolites covaried with at least one other metabolite. The density (see Methods) of this network is 0.15, and six modules (Figure 3A) were found in this network using a network module detection method that optimizes a modularity function through simulated annealing (Guimerà and Nunes Amaral, 2005). The abundance profiles were normalized into vectors with mean zero and variance one and grouped together according to the six modules found in Figure 3A. The abundance change patterns of the metabolites in each module are shown in Figure 3B. Metabolites in one of the two largest modules (Figure 3B, green symbols) showed transient upregulation after 2 min of ABA treatment, and then downregulation after 10 and 30 min of ABA treatment. Interestingly, metabolites in the other large module (Figure 3B, yellow symbols) also showed transient upregulation at 2 min of ABA treatment, and then returned to basal levels at 10 min, followed by another upswing at 60 min. The abundance change patterns of metabolites of the third largest group in Figure 3A showed progressive downregulation after ABA treatment until 30 min, and then showed an increase (Figure 3B, aqua symbols).
Figure 3.
The ABA-Regulated Metabolome of Col Guard Cells Exhibits a Dense Correlation Network with Three Major Temporal Patterns.
The metabolite correlation network and the abundance change patterns of the metabolites in Col that show significant abundance changes (P < 0.01) after treatment with ABA are plotted.
(A) The metabolite correlation network.
(B) Abundance change patterns of the metabolites in each color-coded temporal module.
2-iPMA, 2-isopropylmalic acid; 3-iPMA, 3-isopropylmalic acid; 3PGA, 3-phosphoglyceric acid; ABA-GE, abscisic acid Glc ester; IBA, indole butyric acid; KMBA, methylthiobutyric acid; MeIAA, methyl indole acetate; NDGA, nordihydroguaiaretic acid; OAEE, oleic acid ethylester; OPDA, 12-oxophytodienoic acid.
In gpa1, 43 of the 85 metabolites showed significant abundance changes over time after treatment with ABA and 39 of the 43 metabolites covaried with at least one other metabolite (Figure 4A). The density of the gpa1 metabolic correlation network was 0.13, somewhat less than that of the network in Col. The abundance change patterns of the metabolites are shown in Figure 4B. There is a prominent absence of patterns with peaks at 2 min in the gpa1 mutant when exposed to ABA, which is quite different from Col.
Figure 4.
The ABA-Regulated Metabolome of gpa1 Guard Cells Exhibits a Sparser Correlation Network than in Col and Shows Different Temporal Patterns.
The metabolite correlation network and the abundance change patterns of metabolites in gpa1 that show significant abundance changes (P < 0.01) after treatment with ABA are plotted.
(A) The metabolite correlation network.
(B) Abundance change patterns of the metabolites in each color-coded temporal module.
3PGA, 3-phosphoglyceric acid; DHJA, dihydrojasmonic acid; MeDHJA, dihydrojasmonic acid methylester; NDGA, nordihydro guaiaretic acid; OAEE, Oleic Acid Ethylester; OPDA, 12-oxophytodienoic acid.
Time-Course Profiling Reveals a Differentially Regulated Metabolome in Col versus gpa1
To identify metabolites that show differential abundance over time between ethanol and ABA treatments, or between wild-type and mutant genotypes, we used a statistical approach specifically designed to ask whether two curves (time courses) differ significantly from each other (Storey et al., 2005). This approach is implemented in EDGE software (Storey et al., 2005).
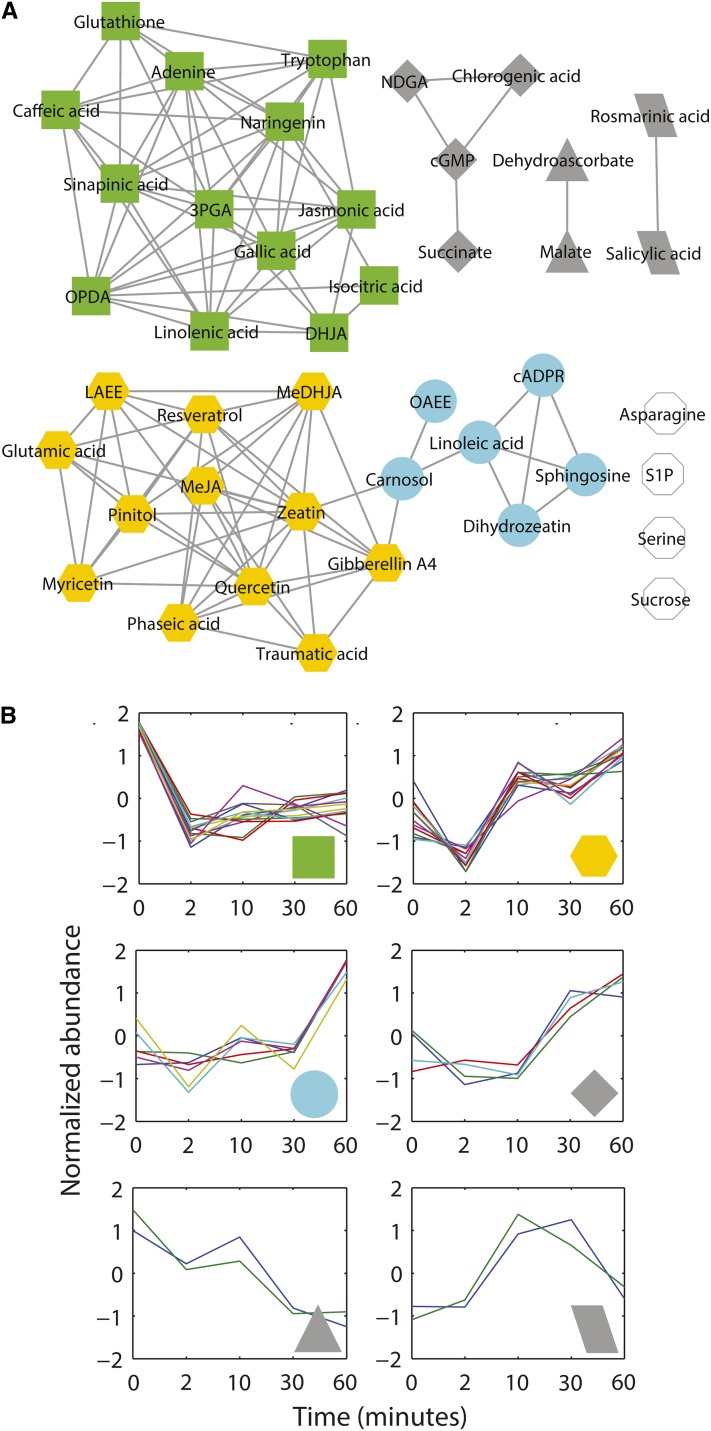
In Col (Figure 5A), 41 metabolites showed significantly different time courses between solvent and ABA conditions (P < 0.05 and Q < 0.05). Many of these metabolites showed an increase at the 2-min time point and then decreased, as shown in the heatmap (Figure 5A) and in the two largest modules of Figure 3A (green and yellow symbols). Nine metabolites—indole-3-acetic acid (IAA), protocatechuic acid, cinnamic acid, Suc, zeatin glucoside, syringic acid, indole carboxylic acid (ICA), jasmonic acid methyl ester (MeJA), and Met—were downregulated by ABA (Figure 3B, aqua symbols). Compared with the ethanol control, only cADPR, jasmonic acid (JA), rosmarinic acid, and oxaloacetic acid showed upregulation by ABA exposure at all time points in Col; the time courses of these metabolites did not covary with each other so they are not connected by edges in Figure 3A.
Figure 5.
Heatmaps of Normalized Abundance of the Metabolites Show Differential Responsiveness of Col and gpa1 Guard Cell Metabolomes to ABA.
(A) The metabolites in Col that show statistically significant temporal differences in abundance between ethanol and ABA treatments (EDGE, P < 0.05 and Q < 0.05 in all cases). The green, yellow, aqua, gray, and white symbols correspond to the modules of Figure 3.
(B) The metabolites in gpa1 that show statistically significant temporal differences in abundance between ethanol and ABA treatments (EDGE, P < 0.05 and Q < 0.05 in all cases). The green, yellow, aqua, gray, and white symbols correspond to the modules of Figure 4.
(C) The metabolites that show statistically significant temporal differences in abundance between Col and gpa1 over a time course of ABA treatment (EDGE, P < 0.05 and Q < 0.05 in all cases).
2-iPMA, 2-isopropylmalic acid; 3-iPMA, 3-isopropylmalic acid; 3PGA, 3-phosphoglyceric acid; ABA-GE, abscisic acid Glc ester; DHJA, dihydrojasmonic acid; IBA, indole butyric acid; KMBA, methylthiobutyric acid; MeIAA, methyl indole acetate; NDGA, nordihydro guaiaretic acid; OAEE, oleic acid ethylester; OPDA, 12-oxophytodienoic acid.
In contrast with the 41 metabolites in Col that showed statistically significant differences in ABA versus ethanol time courses, only 16 metabolites were found to have statistically significant differences in their time courses between solvent control and ABA treatment in gpa1 (Figure 5B). In contrast with Col, these metabolites did not show a peak at 2 min and instead showed either a strong (Figure 5B, green symbols) or moderate (Figure 5B, yellow symbols) decline by 2 min or little change in abundance at that time (Figure 5B, aqua and gray symbols). Several metabolites, including linolenic acid ethylester (LAEE), pinitol, cADPR, sphingosine, carnosol, succinate, and cGMP showed a strong increase at 60 min. When ethanol-treated samples were compared between Col and gpa1 using EDGE software, none of the 85 metabolites satisfied the significance criteria, indicating that these genotypes do not differ significantly in their metabolite time courses after ethanol treatment. However, 57 of the 85 metabolites quantified displayed pronounced differences between Col and gpa1 toward ABA treatments (Figure 5C). We found that many of these metabolites were plant hormone or hormone-related metabolites, including the following: (1) the auxin IAA and the auxin-related compounds indole butyric acid, ICA and methyl indole acetate; (2) JA and the related compounds MeJA and dihydrojasmonic acid, as well as their precursors LAEE, linolenic acid, linoleic acid, and 12-oxophytodienoic acid; (3) the gibberellins GA3 and GA4; (4) the cytokinins zeatin and dihydrozeatin and the glycosylated product zeatin glucoside; and (5) salicylic acid and the related compounds methyl salicylate, protocatechuic acid, and syringic acid. With the notable exceptions of IAA (see below) and indole butyric acid, these hormones were generally lower in concentration in gpa1 than in Col.
Because treatment with ABA-containing solution inevitably resulted in residual ABA solution being collected with the protoplasts, we made no attempt to quantify ABA itself. However, the ABA catabolite dihydrophaseic acid, as well as the inactive ABA Glc ester was generally more abundant in gpa1 than in Col, whereas the reverse was true for the ABA catabolite phaseic acid. Several known ABA signaling metabolites of guard cells also differed in their ABA time courses between the two genotypes, including the Ca2+-mobilizing agents sphingosine-1-phosphate (S1P), inositol 1,4,5 trisphosphate (InsP3), cADPR, and cGMP.
In Supplemental Figure 3 online, a vector diagram showing metabolites that significantly differed (by Student’s t test) at each individual time point in ABA versus ethanol guard cell samples at the same time point further illustrates genotype-specific ABA-responsive metabolites. At 2, 10, 30, and 60 min of ABA exposure, there were 21, 21, 41, and 27 metabolites, respectively, that showed significant ABA response in at least one genotype. Only very few metabolites, however, shared the same response trends in Col and gpa1. Only one, two, five, and two metabolites were similarly regulated by ABA in both Col and gpa1 at 2 min, 10 min, 30 min, and 60 min, respectively. These metabolites were as follows: LAEE (downregulated at 2 min); rosmarinic acid (upregulated at 10 min) and LAEE (downregulated at 10 min); salicylic acid, rosmarinic acid (upregulated at 30 min), Suc, malate, and Ser (downregulated at 30 min); and cADPR (upregulated at 60 min) and Suc (downregulated at 60 min). These results again indicate the importance of GPA1 for ABA regulation of the dynamic guard cell metabolome.
Guard Cell Signaling Molecules
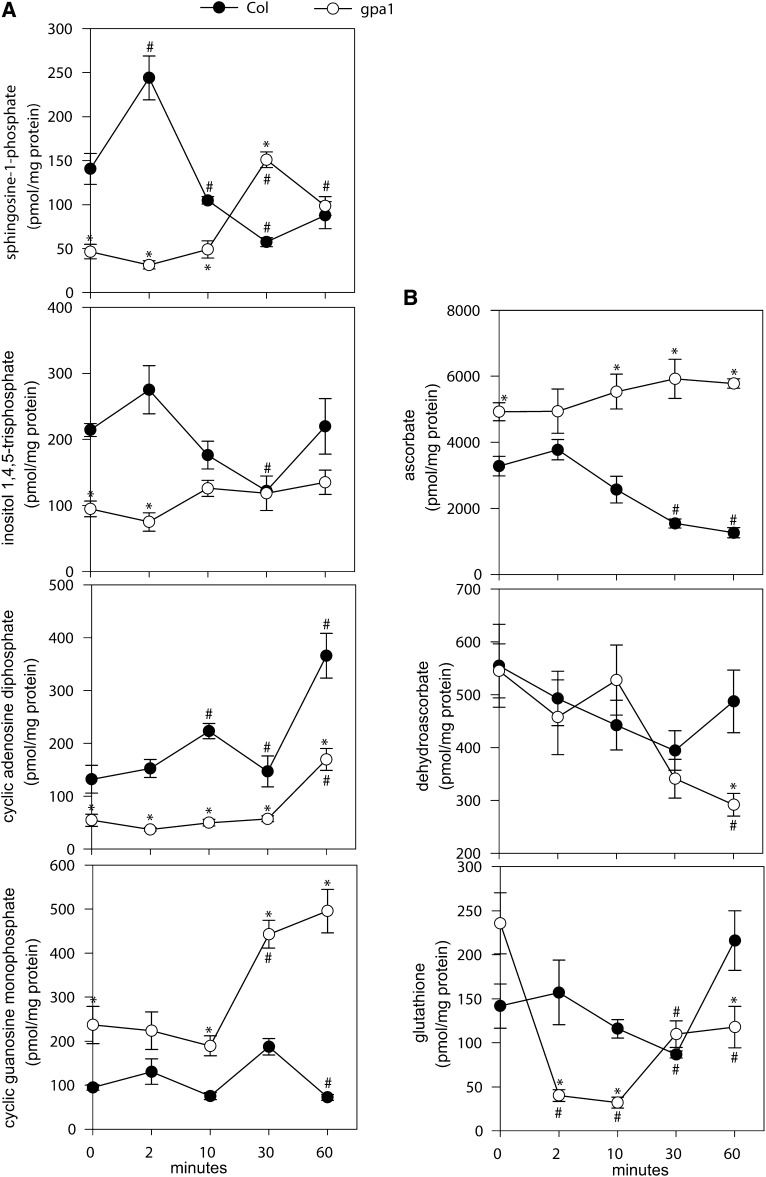
A major goal of our research was to quantify ABA-induced changes in concentrations of known metabolites related to guard cell signaling. Figure 6A shows quantification of four metabolites implicated in Ca2+ mobilization in guard cells. In Col but not gpa1, S1P shows a transient increase at 2 min, consistent with our previous targeted biochemical analyses (Coursol et al., 2003). S1P also shows a significant (Student’s t test) transient increase in gpa1 but this occurs later, at 30 min, and overall S1P levels are lower in gpa1 than in Col. InsP3 similarly showed lower levels in gpa1 than in Col. cADPR showed an increase in Col starting at 10 min, whereas cADPR increased significantly in gpa1 only at the 60-min time point of ABA treatment and also was present overall at a lower concentration in gpa1 than in Col. cGMP showed an ABA-induced increase in gpa1 that was significantly different from the solvent control only at 30 min, whereas this metabolite was lower in Col and was actually slightly downregulated by ABA at 60 min.
Figure 6.
Comparison of Metabolite Time Courses in Col versus gpa1 Guard Cells of Metabolites Known or Implicated in Guard Cell ABA Signaling.
(A) Metabolites implicated in elevation of [Ca2+]cyt.
(B) Metabolites involved in redox regulation.
Error bars indicate the sd (n = 4). Asterisks indicate significant differences between genotypes. Hatch marks indicate that the data point was significantly different within a genotype between ABA treatment and ethanol control (Student’s t test; P < 0.05).
ROS are major players in guard cell signaling and ROS levels will be affected by scavengers such as ascorbate and GSH. We quantified ascorbate (reduced) and dehydroascorbate (oxidized) as well as GSH in our guard cell samples (Figure 6B). Unfortunately, we could detect but not reliably quantify GSSG in our samples because of its low abundance, leading to high variance in quantitation. In Col, ascorbate levels declined after ABA treatment, although this was not accompanied by a statistically significant change in dehydroascorbate (Figure 6B). In gpa1, ascorbate did not change in abundance after ABA treatment, and dehydroascorbate was significantly different (lower) than the parallel ethanol control only at 60 min (Figure 6B). Ascorbate levels were overall greater in gpa1 than in Col. For GSH, in Col, only a slight and transient decrease in GSH levels was observed, whereas in gpa1 GSH was significantly lower than the ethanol control time points at all time points after ABA treatment, and also tended to be lower than the levels found in Col (Figure 6B).
ABA Regulation of Other Hormones
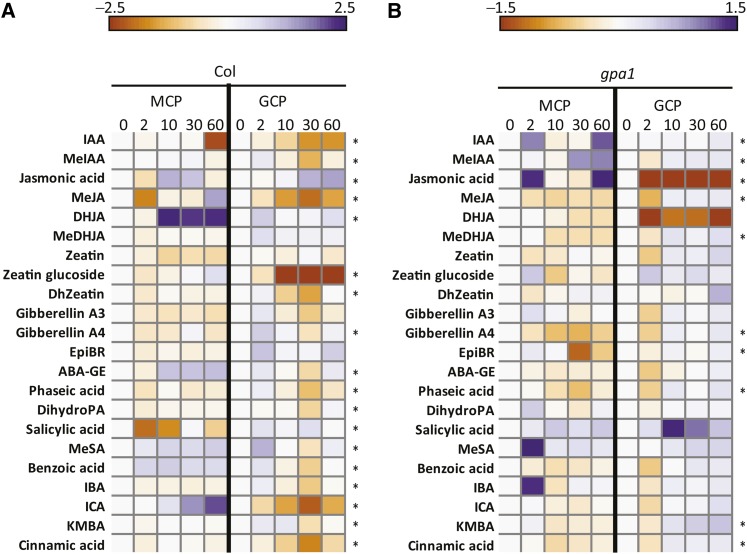
Our analysis of the guard cell metabolome revealed that the abundance of many hormone-related metabolites was modulated by ABA in Col, with this modulation being disrupted in the gpa1 knockout mutants (Figures 5A and 5B). When the same 22 hormones/hormone-related metabolites were measured in mesophyll protoplasts treated with an identical time course of ABA exposure, profiles of 18 of these metabolites in Col and 10 of these metabolites in gpa1 differed in mesophyll cells compared with guard cells (Figures 7A and 7B). These results demonstrate that the guard cell metabolome response to ABA is not a generic plant cell response to ABA. In ethanol-treated mesophyll samples, there are no metabolites in either Col or gpa1 that show significant (P < 0.01) changes in abundance along the time course, confirming that the changes described above were indeed induced specifically by ABA.
Figure 7.
Comparison of Hormone Metabolite Time Courses in Response to ABA in Guard Cell Protoplasts versus Mesophyll Cell Protoplasts of Col and gpa1 Mutants.
(A) Comparison in Col.
(B) Comparison in gpa1.
Heatmaps in this figure represent log ratios, that is, the metabolite level at each time point was divided by the level at 0 min and then subjected to a log2 transformation, to facilitate comparison of temporal changes across the two cell types. Asterisks indicate metabolites with significantly different time courses in response to ABA in guard cells versus mesophyll cells. (EDGE, P < 0.05 and Q < 0.05 in all cases).
ABA-GE, abscisic acid Glc ester; DHJA, dihydrojasmonic acid; DihydroPA, dihydrophaseic acid; DhZeatin, dihydrozeatin; EpiBR, epibrassinolide; GCP, guard cell protoplast; IBA, indole butyric acid; KMBA, methylthiobutyric acid; MCP, mesophyll cell protoplast; MeDHJA, dihydrojasmonic acid methylester; MeIAA, methyl indole acetate; MeSA, methyl salicylate.
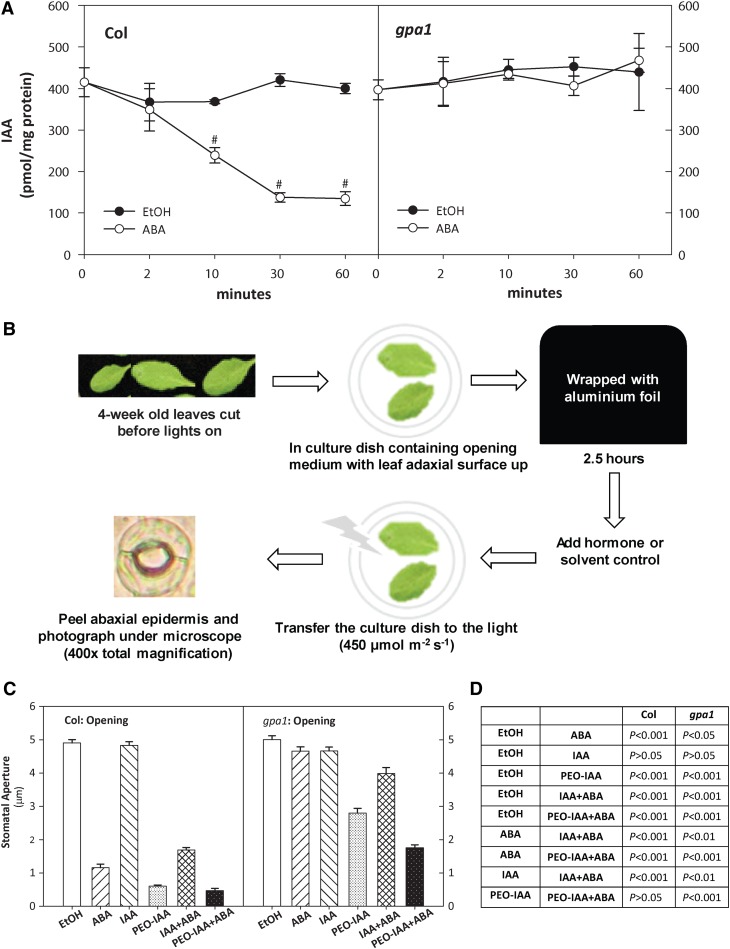
ABA Regulation of IAA in Guard Cells
In particular, ABA induced a significant downregulation of IAA levels in Col but not in gpa1 mutant guard cells (Figure 8A); the same pattern was not seen in mesophyll cells (Figure 7). There is previous literature showing that application of exogenous auxin can enlarge stomatal apertures (Snaith and Mansfield, 1982; Lohse and Hedrich, 1992; She and Song, 2006; Tanaka et al., 2006). We hypothesized that downregulation of endogenous IAA concentration could be a component mechanism for ABA inhibition of stomatal opening and promotion of stomatal closure, and that absence of this component could contribute to the ABA hyposensitivity of gpa1 guard cells. To test our hypothesis, we evaluated stomatal aperture responses (Figure 8B) to ABA, IAA, and the IAA antagonist PEO-IAA (Zhang et al., 2008; Nishimura et al., 2009; Ishida et al., 2010). ABA inhibited wild-type Arabidopsis (Col) stomatal opening in response to white light, whereas the gpa1 mutant showed ABA hyposensitivity to ABA inhibition of stomatal opening (Figures 8C and 8D), consistent with previous reports. In both genotypes, exogenous IAA application did not further enhance white light–induced opening beyond 5 µm (Figures 8C and 8D). In both genotypes, however, the auxin antagonist α-(phenyl ethyl-2-one)-IAA significantly decreased white light–induced opening, suggesting that endogenous auxin may be required for full stomatal opening under white light. PEO-IAA alone was less effective in inhibiting stomatal opening in gpa1 than in Col. The reason for this difference is unclear but perhaps could reflect an ability of gpa1 to maintain IAA levels. In assays of ABA inhibition of stomatal opening, exogenous IAA application partially reversed ABA inhibition of opening in Col but not in gpa1, perhaps because endogenous IAA levels in gpa1 are already maintained in the face of ABA treatment (Figure 8A). The auxin antagonist PEO-IAA increased the efficacy of ABA inhibition of stomatal opening in both genotypes but this occurred to a much greater extent in gpa1 than in Col. Altogether these results are consistent with ABA-stimulated decreases in IAA as a component of ABA inhibition of stomatal opening, and maintenance of high endogenous IAA levels in the face of ABA treatment (Figure 8A) as a component of the ABA hyposensitivity of the gpa1 genotype.
Figure 8.
ABA Regulates IAA Concentrations in Guard Cells and IAA Is Implicated in ABA Inhibition of Stomatal Opening.
(A) ABA treatment rapidly decreases IAA concentrations in Col but not in gpa1 guard cells. Error bars indicate the sd (n = 4). Hatch marks indicate that the data point was significantly different within a genotype between ABA treatment and ethanol control (Student’s t test; P < 0.002).
(B) Schematic of protocol for measurement of stomatal opening responses.
(C) Stomatal opening responses in Col and gpa1 to ABA, IAA, and the IAA antagonist PEO-IAA suggest IAA participation in the Col guard cell ABA response. Error bars indicate the se (n > 125 stomata per sample).
(D) Significance of differences (Student’s t test) in stomatal aperture values seen in (C).
[See online article for color version of this figure.]
DISCUSSION
The Single Cell Type Metabolome of Guard Cells
Cellular level metabolite analysis not only requires highly sensitive instruments, but also facile isolation of large quantities of distinct cell types. Much previous research has confirmed that guard cell protoplasts retain ABA responsiveness such as in ion channel and transcriptome regulation by ABA (Leonhardt et al., 2004; Kim et al., 2010; Pandey et al., 2010; Wang et al., 2011; Hedrich, 2012). We additionally found that, as expected, guard cell protoplast volume declined after ABA (but not after ethanol) treatment, confirming retention of cellular functionality (see Supplemental Figure 2 online).
There are not many examples of time-course analysis on single cell metabolomes of plants. We chose to focus our analysis on quantification of known and hypothesized signaling metabolites in the guard cell ABA response. Our metabolome, consisting of 85 metabolites, is small in comparison to the total number of predicted plant metabolites (Goodacre et al., 2004) but is of the same magnitude as those of other studies on stress-regulated metabolomes. For example, the nontargeted studies by Urano et al. (2009), Rizhsky et al. (2004), Rao et al. (2010), and Oliver et al. (2011) focused to a large extent on primary metabolites and identified ∼150, 52, 148, and 167 metabolites, respectively, whereas the study by Ebert et al. (2010) of nonguard cell epidermal cell types in Arabidopsis identified 117 compounds, most of which were primary metabolites.
Ethanol Does Not Significantly Affect the Metabolome
There is a lack of information about ethanol’s effect on plant cell metabolite concentrations. Therefore, we set up each ABA treatment time point with a parallel ethanol control time point to distinguish valid metabolite responses to ABA from responses to ethanol. PCA analysis showed that ethanol samples from both wild-type Col and gpa1 mutant guard cells clustered together with the zero time point samples that received neither ethanol nor ABA treatment (Figure 2), indicating the lack of a systematic ethanol effect. An ANOVA on metabolite levels over the time course of ethanol treatment further confirmed the absence of significant ethanol effects in guard cells or mesophyll cells. These results suggest that this concentration of ethanol has minor effects on cellular metabolomes. These results are also further evidence that the protoplasts were physiologically stable and healthy during the 1-h treatment period. This information can benefit future studies in allowing a reduction in the numbers of ethanol control samples.
Metabolites Group Together in Temporal Modules
Time-course data (Acharya et al., 2013) provide important information for accurate and predictive modeling of cellular signaling networks, including those of guard cells (Li et al., 2006), because they can be used to impose temporal constraints on pathways within a network. We found that there was temporal modularity in the ABA-regulated metabolome of guard cells, with several different temporal patterns observed (Figures 3 and 4). Our results, showing that many metabolites in Col peak in abundance at 2 min of ABA treatment followed by a decline, suggest that certain metabolites are first responders in the ABA response. Our results are consistent with the known rapid response of stomata to ABA (Assmann and Grantz, 1990; Assmann et al., 2000) and also illustrate the value of obtaining time-course data. For example, had we collected data only at 0 and 60 min, we might have concluded erroneously that many of the metabolites in one of the two most common modules (Figure 3, yellow symbols) did not respond to ABA. By inference, it is also true for our own study that additional information might have been revealed by inclusion of additional time points; this was precluded owing to the need to grow ∼30,000 plants for the current guard cell dataset alone. Because many metabolites returned to baseline by 60 min, future investigations may benefit from a focus on finer temporal divisions within a 1-h time period.
The density of edges found connecting the three major modules of Figure 3A suggests that there is strong coordination in the temporal response of the guard cell metabolome. Metabolites that covaried over time after ABA treatment were not in the same or closely related metabolic pathways (Toubiana et al., 2013). This can be seen by comparing the metabolite biosynthetic relationships depicted in Figure 1 with the modules depicted in Figures 3 and 4. For example, the major temporal modules in Col (Figure 3A, green and yellow symbols) each include diverse hormones, organic acids, and lipid metabolites. We also did not observe that temporal patterns in metabolite abundance paralleled biosynthetic pathway relationships. For example, linoleic acid is biosynthetically downstream of LAEE, but both of these metabolites appear within the same temporal module (Figure 3A, green symbols). In the absence of flux analysis, caution must be exercised in drawing conclusions, but it appears that coexpression analysis for temporal metabolome profiling may not be useful as a tool to elucidate the positions of metabolites within known biosynthetic pathways. For example, if nontargeted analysis is performed, it should not be assumed that unknown peaks that cluster temporally with known peaks represent metabolites in the same metabolic pathways as the known peaks. The metabolite diversity found within our modules implies a coordinated regulation of diverse metabolic pathways in the guard cell response to ABA.
Responses of Known Metabolites in the Guard Cell ABA Signaling Network
Our metabolite analysis was focused on quantitation of known nonprotein secondary messengers of guard cell signaling. The majority of these metabolites have not been measured in other studies of ABA/drought-regulated metabolomes, which have focused on analysis of whole shoots (Urano et al., 2009) or leaves (Rizhsky et al., 2004; Evers et al., 2010; Oliver et al., 2011) with an emphasis on primary metabolites.
Ca2+ Regulation
We obtained temporal profiles for a number of metabolites previously implicated in Ca2+ (Figure 6A) and redox (Figure 6B) regulation of guard cell ABA signaling. The second messengers S1P, InsP3, cADPR, and cGMP have been reported as calcium ion mobilizing agents in plants (Berridge, 1993; Allen et al., 1995; Zhao et al., 2005). Ng et al. (2001) first reported that an elevation in S1P occurs in drought-stressed leaves and that S1P induces [Ca2+]cyt oscillations in guard cells and promotes stomatal closure in Commelina communis. Biochemical assays on Arabidopsis guard cell protoplasts showed an ABA-induced peak in S1P at 2 min (Coursol et al., 2003). However, the role of sphingoid bases in plant signaling has been challenged (Lynch et al., 2009). Our metabolite analyses, showing that S1P levels in Col guard cells peak at 2 min after ABA treatment and then decline (Figures 3B and 6A), verify the results of Coursol et al. (2003) and further implicate phosphorylated sphingoid bases in ABA signaling (Coursol et al., 2003, 2005; Worrall et al., 2008). The observation that insertional mutagenesis of sphingosine kinase1 (At4g21540) in Arabidopsis results in reduced stomatal aperture responses to ABA, whereas overexpression of this enzyme leads to ABA hypersensitivity (Worrall et al., 2008) is consistent with this conclusion.
Experimental elevation of intracellular InsP3 concentrations induces cytosolic Ca2+ elevation and stomatal closure in C. communis (Gilroy et al., 1990) and inhibits K+ uptake channels in Vicia faba (Blatt et al., 1990). Transgenic tobacco (Nicotiana tabacum) with very low levels of phospholipase C shows reduced ABA inhibition of stomatal opening but wild-type stomatal closure responses to ABA (Hunt et al., 2003; Mills et al., 2004), although pharmacological phospholipase C inhibitors do impair ABA-induced Ca2+ oscillations and stomatal closure in C. communis, V. faba, and tobacco (Jacob et al., 1999; Staxén et al., 1999; MacRobbie, 2000; Mills et al., 2004). In our measurements (Figure 6A), InsP3 showed a peak at 10 min of ABA application in Col but this was not statistically different from the ethanol control (Student’s t test, P = 0.427). In the earliest literature on InsP3 measurement in guard cells, wherein InsP3 was quantified using a validated radioreceptor assay, the ABA-induced peak in InsP3 was seen at 10 to 60 s after ABA application to V. faba guard cell protoplasts, followed by a rapid return to baseline (Lee et al., 1996). These results suggest that our first time point, at 2 min, might already have missed the major ABA-induced change in InsP3 levels. The possibility that InsP3 elevation participates in the most initial or rapid aspects of ABA signaling also could be consistent with the observation that short-term inhibitor experiments show greater effects on stomatal closure than long-term genetic manipulation of InsP3 metabolism (Mills et al., 2004). It is also possible that phosphoinositide metabolites other than InsP3 may be more central to ABA signaling, and that changes in their concentration may require altered flux through phosphoinositide pathways that do not necessarily alter concentrations of InsP3. For example, InsP6 (Lemtiri-Chlieh et al., 2000; Lemtiri-Chlieh et al., 2003) as well as phosphatidylinositolphosphate (PIP), PIP2, PIP4, and PIP5 all have been implicated in various aspects of guard cell function (Lee et al., 1996; Jung et al., 2002; Lee et al., 2007). A comprehensive test of this hypothesis would require detailed flux analysis of phosphoinositide metabolites in guard cells.
Intracellular application of cADPR in guard cells elicits cytosolic Ca2+ oscillations (Leckie et al., 1998; Garcia-Mata et al., 2003) in C. communis and V. faba. Such exogenous cADPR application also promotes stomatal closure, whereas cADPR antagonists interfere with ABA-induced stomatal closure in V. faba (Leckie et al., 1998). These experiments led us to measure endogenous cADPR. Our results (Figures 5C and 6A) show a gradual increase in cADPR levels in Col guard cells that is not seen in gpa1, followed by a more marked increase in Col and a smaller increase in gpa1 at 60 min. The time courses suggest that cADPR may not only participate in short-term signaling but may also play a role in longer-term guard cell responses to ABA and drought.
cGMP or a derivative has been implicated (using pharmacological agents) as a secondary messenger that operates downstream of ROS and NO and upstream of Ca2+ elevation in ABA-induced stomatal closure (Neill et al., 2002; Dubovskaya et al., 2011). cGMP increases in concentration in whole Arabidopsis seedlings at 60 s after ABA application (Dubovskaya et al., 2011). By contrast, cGMP in Col guard cells did not increase upon ABA treatment, whereas a delayed increase was found in gpa1 (Figure 6A; see further discussion below).
Redox Regulation
Many enzymes and metabolites contribute to cellular redox regulation and only a subset of these was quantified in this initial guard cell metabolome, so our data should be interpreted with caution. The observations in Col (Figure 6B) that ascorbate declined but dehydroascorbate did not increase with ABA exposure, whereas changes in GSH levels were largely nonsignificant, suggest that other antioxidants are utilized in guard cell ABA signaling, that there is flux through the ascorbate-GSH cycle but little change in standing metabolite concentrations, or that changes in these metabolites occurred at time points other than those included in our time course.
ABA-Regulated Metabolomes Are Altered in gpa1: Metabolome Modulation by the Heterotrimeric G Protein
Elucidation of the role of heterotrimeric G proteins in hormonal signaling, stress regulation, and plant development has advanced greatly over the past decade (Wang et al., 2001; Jones et al., 2003; Perfus-Barbeoch et al., 2004; Assmann, 2005, 2010; Joo et al., 2005; Pandey et al., 2006; Tsugama et al., 2013; Urano et al., 2013). Previous studies revealed that the G-protein α subunit mutant gpa1 is hyposensitive to ABA inhibition of stomatal opening but exhibits wild-type sensitivity to ABA promotion of stomatal closure (Wang et al., 2001), and shows hyposensitivity in ABA inhibition of inward K+ channels, ABA activation of Ca2+-permeable channels, and pH-independent activation of slow anion efflux channels, as well as in ABA stimulation of ROS production (Wang et al., 2001; Fan et al., 2008; Pandey et al., 2010; Wang et al., 2011; Zhang et al., 2011). Our metabolome analyses (Figures 2 to 8) demonstrate that there is also ABA hyposensitivity of the gpa1 guard cell metabolome.
PCA initially revealed that the ABA-treated metabolomes of wild-type Col guard cells clearly separate from those of gpa1 samples in PCA (Figure 2). ABA-treated Col samples displayed more divergence from their control samples than did gpa1 mutant samples from their controls (Figure 2), consistent with ABA hyposensitivity of gpa1. EDGE analysis similarly revealed that 41 of 85 metabolites had significantly different ABA versus ethanol time courses, whereas this was true for only 16 metabolites in gpa1 (Figure 5).
Fewer network modules and lower network density were observed in the ABA-treated gpa1 metabolome compared with that of Col (Figures 3 and 4). The reduced connectivity of the gpa1 metabolome (Figures 3 and 4) suggests that loss of GPA1 disrupts coordination of the cellular response to ABA. A frequent question concerning Arabidopsis G protein–related phenotypes has been how the single Gα subunit encoded in the Arabidopsis genome could affect so many different hormonal and signaling processes. The reduction in module number and intramodule and intermodule connectivity seen in gpa1 versus Col is consistent with the hypothesis (Assmann, 2004, 2005) that GPA1 functions as a modulator of or rheostat on numerous signaling pathways, as opposed to the conventional idea that specific G proteins function in specific linear pathways, as has arisen from the paradigm in mammalian systems, wherein pathway specificity can readily be attained by combinatorial association of numerous Gα, Gβ, and Gγ subunits.
In addition to GPA1 knockout reducing the number of metabolites that respond to ABA in guard cells, the temporal responses of the metabolites that do respond is also altered in this mutant. As can be seen from PCA, ANOVA, and EDGE analyses (Figures 2 to 5), there is a prominent absence of patterns with peaks at 2 min in the gpa1 mutant when exposed to ABA, which is quite different from Col. Moreover, in contrast with Col, many metabolites in the gpa1 mutant had concentrations at 10 and 30 min that were similar to or greater than their zero time point values compared with patterns in Figure 4B. A delay in the response of specific metabolites to ABA could cause ABA hyposensitivity by disrupting time-dependent information flow through the guard cell ABA signaling network, and/or result in slowed guard cell responses to ABA. To date, stomatal aperture and gas exchange analysis of the gpa1 mutant have focused on steady state measurements; the kinetics of stomatal response could be an interesting topic for future investigation (Acharya et al., 2013).
Key Metabolites Differ in Col versus gpa1 Guard Cell Metabolomes
In addition to overall changes in metabolome profiles between Col and gpa1, we were interested in asking which specific metabolites might be more mechanistically relevant to the altered stomatal aperture phenotypes of gpa1. A comparison of Ca2+ regulatory metabolites and regulators of redox status from our metabolomes identifies some individual metabolites that may be of particular interest for future targeted studies.
Regulators of Ca2+ Mobilization and Redox Status
Coursol et al. (2003) observed that the gpa1 mutant was hyposensitive to S1P regulation of stomatal movements and to S1P inhibition of inward K+ channels and activation of slow anion channels (Coursol et al., 2003), whereas Ng et al. (2001) showed that S1P elevates cytosolic Ca2+ in wild-type guard cells. In gpa1 mutants, inward K+ channels are ABA insensitive, whereas slow anion channels are regulated by redundant parallel pathways, one of which requires GPA1 (Wang et al., 2001; Fan et al., 2008). Our metabolite results show a delayed and diminished response of S1P levels to ABA in gpa1. Because K+ channels are inhibited by [Ca2+]cyt, whereas anion channels are activated by [Ca2+]cyt, our results suggest that loss of S1P-based Ca2+ signaling in gpa1 could be central to the modified ABA sensitivity of these channels in this mutant.
InsP3 levels were overall higher in Col than in gpa1. This result is consistent with the idea that InsP3 may primarily be employed as a secondary messenger for ABA inhibition of stomatal opening, for which gpa1 is hyposensitive, as opposed to a role in ABA-induced stomatal closure. Consistent with this hypothesis, transgenic expression in Arabidopsis of mammalian type I inositol polyphosphate 5-phosphatase with concomitant reduction in soluble phosphoinositides results in ABA hyposensitivity of stomatal opening but not stomatal closure (Perera et al., 2008). The gpa1 mutant also exhibited constitutively lower levels of cADPR, which mobilizes Ca2+ from the plant vacuole (Allen et al., 1995) and endoplasmic reticulum (Navazio et al., 2001). Among the four Ca2+-mobilizing agents that we quantified (Figure 6A), cGMP, which has been implicated in ABA promotion of stomatal closure (Neill et al., 2002; Dubovskaya et al., 2011), gave results between genotypes that were initially unexpected in that ABA-hyposensitive gpa1 showed higher cGMP levels than Col and cGMP elevation at later time points after ABA treatment. However, it was recently shown that the form of cGMP active in stomatal closure is 8-nitro-cGMP (Joudoi et al., 2013), whereas exogenous cGMP promotes stomatal opening (Cousson and Vavasseur, 1998; Cousson, 2003; Joudoi et al., 2013); our results are consistent with these observations.
With regard to redox signaling, ABA downregulated ascorbate levels in Col (Figures 3 and 6B) but not in gpa1, in which ascorbate pool size was higher than in Col (Figure 6B). GSH levels decreased dramatically in ABA-treated gpa1 guard cells (Figures 4 and 6B) and were significantly lower than in wild-type Col with ABA treatment (Figures 5C and 6B). Phenolic acids and flavonoids play important roles in plant development and defense, acting as reducing agents and free radical scavengers (Hichri et al., 2011; Kusano et al., 2011). Most phenolic acids and flavonoids in the gpa1 mutant displayed lower levels than in Col when treated with ABA (Figure 5C). It is possible that the observed low levels of phenolics, most flavonoids, and GSH in gpa1 versus Col in the presence of ABA (Figure 5C) reflects efficient removal of ROS through consumption of these antioxidants; this hypothesis could provide a mechanistic explanation for previous findings that gpa1 mutant guard cells show little ROS production in response to ABA (Zhang et al., 2011) and ozone (Joo et al., 2005).
ABA Affects the Concentrations of Multiple Hormones
Although ABA plays an over-riding role in limiting stomatal apertures, evidence has been uncovered for the participation of other hormones in stomatal aperture regulation and for crosstalk between and among these hormones (Merritt et al., 2001; Tanaka et al., 2006; Acharya and Assmann, 2009). A common scenario to explain such crosstalk has been that hormones converge onto shared downstream signaling components; for example, convergence onto the second messengers NO and ROS of ABA and MeJA as positive regulators and auxins and cytokinins as negative regulators (She and Song, 2006; Song et al., 2006; Munemasa et al., 2011) (Figure 9A). Our results suggest the alternative hypothesis that it is not (simply or solely) that concentrations of these hormones are ABA independent with multiple hormones then converging onto signaling components that are shared with ABA, but rather that ABA operates far upstream within the guard cell to regulate the concentrations of the other hormones (Figure 9B). At the whole plant level, a role for drought in regulation of cytokinin synthesis has already been demonstrated (Nishiyama et al., 2011), although the regulation (Figure 5A) is cell autonomous in our system (isolated guard cell protoplasts). Our multihormone hypothesis is supported by the observation in our dataset that the 45 metabolites significantly changed by ABA in wild-type Col guard cells include not only ABA catabolites such as ABA Glc ester, phosphatidic acid, and dihydrophaseic acid, but also IAA, JA, MeJA, and methyl salicylate (Figure 5A). We also found that ∼20 of the 57 metabolites that differed significantly in their time courses between Col and the ABA-hyposensitive genotype gpa1 were hormones and related metabolites (Figure 5C), and, consistent with this observation, that such metabolites contributed strongly to the PCA loadings that distinguished Col from gpa1 (see Supplemental Data Set 2 online). It would have been difficult to envision such a hypothesis without the ability to simultaneously quantify multiple metabolites in one and the same sample, thus illustrating the power of metabolomics approaches for the study of cell signaling.
Figure 9.
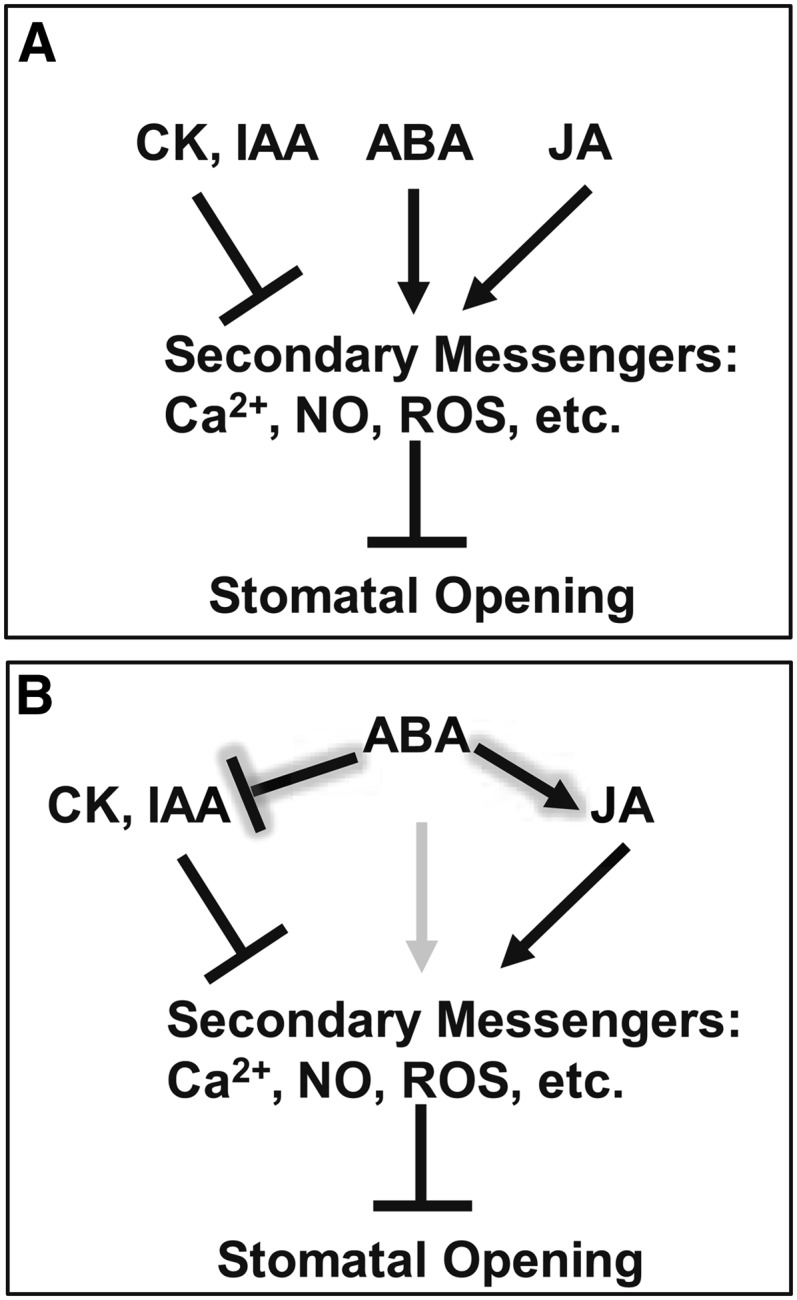
Simplified Models of Hormonal Crosstalk in the Guard Cell ABA Response.
(A) ABA does not regulate concentrations of other hormones. Hormones independently converge on shared downstream signaling elements and crosstalk occurs at the downstream level.
(B) Multihormone model. ABA operates upstream of other hormones to regulate their concentrations.
In both (A) and (B), regular arrows indicate increase or activation; blunted arrows indicate decrease or inhibition. In (B), the arrow indicating a direct effect of ABA on secondary messengers is grayed out, to indicate that some of the apparent direct effects of ABA actually may be mediated through ABA regulation of other hormones. In both panels, CK represents cytokinins, IAA represents indole acetic acid and related auxins, and JA represents jasmonic acid and related compounds.
For an initial evaluation of our multihormone hypothesis, we pursued two approaches. First, we evaluated the hormonal responses of mesophyll cell protoplasts to the identical ABA treatment. We did find changes in hormone concentrations in mesophyll cells in response to ABA; however, the hormonal responses induced in guard cells by ABA were not mirrored in mesophyll cell protoplasts (Figure 7), consistent with the hypothesis that these changes are related to the specialized functions of guard cells.
Second, we specifically assessed the interplay between ABA and IAA in guard cell signaling based on hints of a relationship between ABA and IAA from older reports in the guard cell literature. For example, it was reported that IAA increased stomatal aperture in V. faba (Lohse and Hedrich, 1992; She and Song, 2006), and auxin was found to inhibit ABA-induced stomatal closure in C. communis (Snaith and Mansfield, 1982) and Arabidopsis (Tanaka et al., 2006). It was also of interest that cGMP was previously implicated in IAA induction of stomatal opening (Cousson and Vavasseur, 1998; Cousson, 2003), given that we observed ABA-induced increases in cGMP in the guard cell metabolome of the ABA-hyposensitive gpa1 mutant.
Our metabolome analysis revealed that IAA levels in Col guard cells were reduced significantly starting at 10 min after ABA exposure, whereas no reduction in IAA level was observed in gpa1 guard cells treated with ABA (Figures 5C and 8A). In our assays of stomatal aperture responses (Figure 8B), we found that exogenous IAA (5 µM) opposed ABA inhibition of stomatal opening in wild-type Col but not in the gpa1 mutant (Figures 8C and 8D), consistent with the observed maintenance of IAA levels in the mutant after ABA treatment (Figure 8A). The auxin antagonist PEO-IAA inhibited stomatal opening in both genotypes (Figures 8C and 8D), suggesting that endogenous auxin may be required for full stomatal opening under white light; this would be consistent with the well-documented auxin stimulation of H+ ATPase activity (Lohse and Hedrich, 1992; Takahashi et al., 2012). PEO-IAA partially restored ABA sensitivity of stomatal opening in gpa1 (Figures 8C and 8D), indicating that maintenance of high IAA levels in gpa1 guard cells after ABA treatment contributes to ABA hyposensitivity of stomatal aperture regulation in gpa1. PEO-IAA also enhanced the efficacy of ABA in the wild type (Figures 8C and 8D). These data are consistent with the ABA regulation of IAA levels observed in our metabolome analysis. Accordingly, our data support a multihormone model of ABA function in guard cells (Figure 9B).
In the future, additional experiments on other hormones will allow further assessment of the multihormone hypothesis. Jasmonates may be of particular interest in this regard. In Col guard cells, the JA biosynthesis pathway metabolites linoleic acid, linolenic acid, LAEE, MeJA, and JA all were regulated by ABA (Figure 5A). In particular, JA was strongly induced by ABA at later time points in Col but these responses were not seen in gpa1 (Figure 5C). It has been documented that JA to some extent mimics ABA in accumulating under drought and salt stresses (Creelman and Mullet, 1997; Arbona et al., 2010) and eliciting stomatal closure (Gehring et al., 1997; Suhita et al., 2003, 2004; Munemasa et al., 2007; Hossain et al., 2011) and ABA is required for stomatal responsiveness to JA in tomato (Herde et al., 1997) and Arabidopsis (Hossain et al., 2011), but a role for ABA in regulating the accumulation of JAs pathway metabolites in guard cells has not been previously studied.
A major focus of metabolomics to date has been on elucidation of the flow of mass during substrate to product conversion in one or more biochemical pathways. By contrast, our analysis of the guard cell metabolome highlights the utility of metabolomics as a tool for the elucidation of the flow of information in cellular signaling networks. On the basis of our results, new components can be added to models of guard cell ABA signaling. Our time-course data also can be used to impose temporal constraints on changes in known signaling metabolites. Both types of information should result in improved accuracy of guard cell ABA intracellular signaling models (Li et al., 2006). Identification of the metabolome is also a necessary component of integrated omics analyses. For example, ABA hyposensitivity is more evident in the guard cell gpa1 metabolome than in the guard cell gpa1 transcriptome (Pandey et al., 2010). Among 43 metabolites that have different ABA responses in Col and gpa1 (Figures 5A and 5B), there are 29 metabolites that are nonresponsive to ABA in gpa1, leading to at least 67% metabolites that are ABA hyposensitive in gpa1, a percentage much higher than the 21% ABA-hyposensitive genes in the gpa1 guard cell transcriptome (Pandey et al., 2010). This may reflect the fact that heterotrimeric G proteins are more commonly associated with relatively rapid signaling events that are less likely to include transcriptional regulation.
Several caveats apply to our dataset. First, additional focused analyses on key hormones will be required in the future to further evaluate our multihormone hypothesis of ABA signaling. Second, a more detailed time course, although operationally precluded here by the additional millions of guard cell protoplasts it would require, may have revealed significant changes in metabolite concentrations that were missed by our sampling scheme. Third, it is important to note that metabolites that did not change in concentration could still have experienced treatment- and/or genotype-dependent alterations in their synthesis and catabolism, without a net change in their standing concentration: metabolic flux analysis (Allen et al., 2009) would be required to address this issue. Fourth, although our study provides a large targeted and temporal metabolite analysis of a specific plant cell type, only a minority of all metabolites were quantified. Given the chemical diversity of the metabolome, it will be some time before high-throughput tools are available to identify and quantitatively characterize the metabolome as completely as is currently possible for the transcriptome or even the proteome. Nevertheless, despite their incompleteness, metabolite analyses such as ours demonstrate the power of metabolomics to raise new hypotheses and refine extant models of cellular biochemical and signaling pathways.
METHODS
Plant Growth, Guard Cell Protoplast and Mesophyll Cell Protoplast Isolation, and ABA Treatments
The Arabidopsis thaliana heterotrimeric G-protein α subunit mutants gpa1-3 and gpa1-4 are previously characterized protein null mutants in the Col-0 background that have been used extensively in analyses of guard cell function (Wang et al., 2001; Jones et al., 2003; Perfus-Barbeoch et al., 2004; Joo et al., 2005; Pandey et al., 2006; Fan et al., 2008; Nilson and Assmann, 2010; Zhao et al., 2010). Seeds from these genotypes were sterilized and plated on growth medium containing one-half Murashige and Skoog basal mixture (Sigma-Aldrich), 1% (w/v) Suc, and 0.8% (w/v) plant cell culture tested agar (Sigma-Aldrich) in 150-mm plates. After 2 d of stratification (4°C) in darkness, the plates were transferred to a controlled-environment growth chamber under 120 µmol m−2 s−1 light, with 8-h/16-h day/night cycles and 22°C/20°C day/night temperature cycles for 10 d. Healthy and uniform seedlings were then transplanted to potting mix (Miracle-Gro Inc.) in 5 × 4.5-inch square pots and were watered twice weekly with deionized water. Healthy and young fully expanded rosette leaves from 4- to 5-week-old plants were the tissue source for guard cell protoplast and mesophyll cell protoplast isolation.
Large-scale guard cell protoplast isolations were performed using the two-step enzyme digestion method (Pandey et al., 2002) with some modifications as described below. The guard cell protoplast preparation method was scaled up to 320 leaves per isolation. Basic solution as in Pandey et al. (2002) except without ascorbic acid was used but enzyme compositions were changed. For each isolation, the blended peels were digested in 350 mL enzyme solution 1 containing 0.4% Cellulysin cellulase, Trichoderma viride (Calbiochem), and 0.3% Cellulase R-10, for 1 h. Digestion was continued in 350 mL enzyme solution 2 containing 1% (w/v) Onozuka RS cellulase (Yakult Honsha), and 0.002% (w/v) Pectolyase Y-23 (Seishin Pharmaceutical) for 45 min. After purification on a Histopaque (Sigma-Aldrich) cushion, the guard cell protoplasts were resuspended in basic solution, and washed twice by centrifugation at 150g to remove the enzyme solution. To obtain enough homogenous guard cell protoplasts for metabolomics analysis, we performed three isolations simultaneously from one genotype and suspended guard cell protoplasts from these three isolations in 13.6 mL basic solution. After leaving the guard cell protoplasts on ice in darkness for 1 h, 10 μL of the guard cell protoplast suspension were used for estimation of yield and purity by counting cell number using a hemocytometer (Hausser Scientific), during which time the remaining guard cell protoplasts were left under room light at room temperature for ∼20 min. Any guard cell protoplast isolations with >2% mesophyll cell protoplast contamination, as quantified by observing under the microscope and counting numbers of guard cell protoplasts versus mesophyll cell protoplasts, were discarded.
The three guard cell protoplast isolations were combined and then aliquots were taken for each of the nine treatments. Aliquots (1.5 mL) of guard cell protoplasts were treated with 1.5 μL ABA (50 mM stock, final concentration is 50 µM ABA; A.G. Scientific) or ethanol (solvent control) for 0 (no ABA or ethanol), 2, 10, 30, or 60 min, respectively. At the end of the treatment, guard cell protoplasts were spun down at 150g for 5 min at 4°C and supernatants were removed as much as possible. Each guard cell protoplast pellet was flash-frozen with liquid nitrogen in a 1.5-mL tube.
Mesophyll cell protoplasts were isolated as previously described (Pandey et al., 2002). Briefly, leaves were excised and immersed in distilled water before processing. The abaxial epidermis of each leaf was peeled off using forceps and the mesophyll layer was placed facing down into the enzyme solution (0.5% Macerozyme R-10, 1% Onozuka RS cellulase, 0.01% pectolyase Y-23, 0.25% BSA, 0.1% polyvinylpyrrolidone-40, w/v). After gentle shaking for 1.5 to 2 h, the released mesophyll cell protoplasts were collected by centrifugation at 150g for 5 min and resuspended in 5 mL solution containing 10 mM MES-KOH, pH 5.5, and 0.6 M Suc. Five milliliters of basic solution (10 mM MES-KOH, 0.6 M sorbitol, 0.5 mM CaCl2, 0.5 mM MgCl2, 0.5 mM ascorbic acid, 10 µM KH2PO4, pH 5.5) was layered onto the resuspended cell solution, followed by centrifugation for 5 min at 150g. The interface, containing intact mesophyll cell protoplasts, was transferred to a new tube. Basic solution was added to remove Suc by centrifugation for 5 min at 150g. The final pellet was resuspended in basic solution and kept on ice in darkness for 1 h. Protoplasting yield was estimated by counting cell numbers using a hemocytometer (Hausser Scientific). After placement under room light at room temperature for 20 min, aliquots (999 µL) of mesophyll cell protoplasts were treated with ABA or ethanol identically as for guard cell protoplasts. The mesophyll protoplasts were then collected by centrifugation at 150g for 5 min at 4°C and the supernatants were removed as much as possible. Each sample was then flash-frozen with liquid nitrogen and stored in a −80°C freezer.
Guard Cell Protoplast and Mesophyll Cell Protoplast Protein Concentration Assay
Before metabolite extraction, tubes of stockpiled guard cell protoplasts from the same genotype and treatment were combined as needed to yield sufficient guard cell protoplasts for one replicate. Protein content for each combined sample was determined using the Bradford method (Bradford, 1976). One μL guard cell protoplast or mesophyll cell protoplast sample was added to 799 μL extraction buffer (0.0075% TritonX-100). After vortexing, the solution was left at room temperature for 10 min and then 200 μL of Bio-Rad Bradford protein assay reagent (Bio-Rad) were added. After incubation at room temperature for 5 min, absorbance at 595 nm was measured using a Beckman DU-7500 UV/VIS spectrophotometer. Three replicate measurements were made for each sample and averaged. BSA standard from G-Biosciences (Cat#786-006) was used to calibrate and quantify the samples. Protein content for each sample of guard cell protoplasts and mesophyll cell protoplasts is presented in Supplemental Data Set 3 online.
Metabolite Extraction
For metabolite extraction, each combined guard cell protoplast sample or mesophyll protoplast sample was transferred to a 2.0-mL tube that had been preloaded with a methanol-precleaned stainless steel ball (Hartford Technologies). Tubes were placed into a prechilled 48-well tube holder and then loaded onto a 2000 Geno/Grinder machine (Spex/CertiPrep) and shaken for 2 min at a rate of 1900 strokes/min. The homogenized samples were extracted in 0.4 mL cold chloroform:methanol:water (1:3:1) extraction solvent with vortexing for 15 min in a 4°C room in the dark and then sonicated for 5 min in an ice bath. The samples were centrifuged at 13,000g for 15 min at 4°C and then the supernatants were collected. The extraction was repeated four times, yielding a total extraction volume per sample of 1.6 mL. The extracts were vacuum-dried. Extracts were resuspended in 50% methanol and then immediately loaded onto the HPLC autosampler at 4°C.
Four replicates were prepared for Col-0 and two biological replicates were prepared for each of gpa1-3 and gpa1-4. Evaluation of the quality of the replicates by calculation of pair-wise PCCs over all metabolites yielded high PCCs, with most greater than 0.9 (see Supplemental Data Set 4 online).
HPLC-ESI-MS/MS
HPLC-ESI-MS/MS followed the protocol described by Pan et al. (2008) with some modifications. The standards and samples were separated using a Shimadzu HPLC system (LC-20AD/Prominence Liquid Chromatography; DGU-20A5/ Prominence Degasser; SIC-20AC/ Prominence Autosampler; FCV-11AL/solvent Selection Valve unit; LC-10ADvp/Pump) which was coupled to a hybrid quadrupole/linear ion trap mass spectrometer (AB Sciex 4000 QTRAP) equipped with a Turbo Ion Spray (TIS) interface. We optimized the precursor-product ion transitions with best collision energy and declustering potential for each metabolite using authentic standards (see Supplemental Data Set 1 online) in a positive ion mode as [M+H]+. HPLC was performed using a Gemini 5u C18 110Å 250 mm × 4.6 mm column (Phenomenex) at room temperature with mobile phase comprising 0.1% aqueous formic acid (solvent A) and 0.1% formic acid in acetonitrile (solvent B) and the eluates were monitored by MRM. Solvent gradients were optimized for each target metabolite using two different solvent gradients. Solvent gradient method 1 was conducted at a flow rate of 0.5 mL/min: 5% to 80% solvent B for 25 min, and column re-equilibration for 5 min. Solvent gradient method 2 was performed at a flow rate of 0.25 mL/min: 0% to 6% solvent B for 5 min, 6% to 20% solvent B for 10 min, and column re-equilibration for 5 min. ESI parameters were as follows: curtain gas, 30 psi; nebulizer gas (GS1), 50 psi; auxiliary gas (GS2), 55 psi; ion source, 5000 V; and TIS probe temperature, 350°C. Nitrogen was used as the collision gas. The authentic standards were mixed together with a series of concentrations (0.5 to 60 pmol) to obtain the standard curves (see Supplemental Data Set 1 online), which were used to derive absolute quantitation of the corresponding metabolites in the guard cell and mesophyll cell protoplast samples. All metabolite concentrations from the protoplast samples were within the linear ranges of the standard curves. For each cell type, samples were randomized within the HPLC-ESI-MS/MS run.
Stomatal Aperture Measurement
Stomatal aperture measurements were conducted as previously described (Zhao et al., 2008). For stomatal opening assays (Figures 8B to 8D), excised leaves were put in the dark for 2 h to initially close stomata. For the dark treatment, the leaves were placed in 5 mL of the same medium as would subsequently be used during light stimulated stomatal opening. This opening medium consists of 10 mM KCl, 7.5 mM iminodiacetic acid, 10 mM 2-(N-morpholino)-ethane-sulfonic acid–KOH, pH 6.15. The dishes containing the leaves were then transferred to light (450 μmol m−2 s−1; room temperature) and treatments were added as follows: solvent control (ethanol), 50 µM ABA, 5 µM IAA, or 20 µM PEO-IAA (all hormones were prepared as stocks in ethanol). Leaves were incubated for 2.5 h before measurement. Images of abaxial epidermal peels were taken at ×400 total magnification using a Nikon DIPHOT 300 microscope connected to a Nikon E990 camera. At least 125 stomatal apertures were measured per replicate and values are means ± se from at least three independent replicates. All stomatal apertures were measured using free access ImageJ software (version 1.39u). Treatments were blinded during image acquisition and analysis.
Fluorescein Diacetate Vital Staining and Measurement of Guard Cell Diameter in Response to ABA
To assess the viability of Arabidopsis guard cell protoplasts, guard cell protoplasts were stained with fluorescein diacetate (FDA) at a final concentration of 5 μg/ml for 3 min. Guard cell protoplasts stained with FDA were observed with a laser scanning microscope (Carl Zeiss LSM 510 Metasystem) using a C-Apochromat 40×/1.2 W Corr water immersion objective. FDA was excited by the 488-nm line of argon laser at 4.9 A tube current and 94% attenuation (equal to 6% laser intensity) and detected using a bandpass emission filter (500 to 550 nm). Images of guard cell protoplasts were taken at 0, 10, 30, and 60 min after ABA or ethanol treatment. The diameters of guard cell protoplasts were measured from bright-field images using ZEN Lite 2012 software (Blue Edition; Carl Zeiss).
Statistical Analyses
Four replicates for each time point and treatment were obtained for wild-type plants and two replicates were obtained for each of two gpa1 alleles (i.e., gpa1-3 and gpa1-4 mutants). We combined data from gpa1-3 and gpa1-4 alleles for all statistical analyses, following a previously described method (Nilson and Assmann, 2010). Combining alleles increased the biological validity of the experiment: If the two mutant alleles of gpa1 showed divergent responses (because of a background mutation in one gpa1 line and not the other), statistically significant differences would likely not have been detected because of large variance upon combining the genotypes. This strategy (Nilson and Assmann, 2010) also decreased the total number of genotype × treatment combinations and thus allowed us to reduce the number of statistical tests and therefore the inflation of type 1 error. Post hoc, we found that when we compared gpa1-3 and gpa1-4 guard cell time courses by EDGE analysis (see Supplemental Data Set 5 online), none of the metabolites in either ABA or ethanol treatments differed significantly between gpa1-3 and gpa1-4, suggesting that these two genotypes were indeed highly identical, consistent with the results of other analyses (Coursol et al., 2003; Coursol et al., 2005; Pandey et al., 2006; Fan et al., 2008; Nilson and Assmann, 2010), and thus that differences observed between gpa1 and Col could reliably be attributed to the knockout of gpa1 in each gpa1 line.
PCA
To obtain the overall clustering of the samples, we performed PCA (Abdi and Williams, 2010) using Matlab 7.6 software (The Mathworks Inc.). The metabolite abundances were first averaged over replicates, leading to average abundances in a total of 18 guard cell samples (one zero time point plus four successive time points for each of two treatments, in each of two genotypes). The averaged abundances were standardized over samples, such that the overall mean was zero and the variance was one. Individual samples were visualized using a principal component score plot. The metabolites responsible for the differences between samples can be extracted from the corresponding loadings (see Supplemental Data Set 2 online).
Analysis within Genotypes
To check whether the abundance of each metabolite within a genotype changed significantly over time (P < 0.01) after treatment with ethanol or with ABA, one-way ANOVA (Sahai and Agell, 2000) was performed using Matlab 7.6 on the log2-transformed metabolite abundance in each genotype and treatment. The log2 transformation was used to better meet the normality assumption of ANOVA. A pair-wise PCC was then calculated among metabolite profiles that changed significantly over time in response to ABA. Separate PCC analyses were performed for the wild type (Col) and for gpa1 mutants.
Metabolite Correlation Networks
We created a metabolite correlation network for each genotype (Col or gpa1) with ABA treatment data. An edge connecting two metabolites is drawn if their PCC is larger than 0.9. The Cytoscape software package (http://www.cytoscape.org/) was used to visualize the metabolite correlation networks. The density (D) of a network is calculated by D = 2E/N (N − 1), where E is the number of edges and N is the number of nodes in the network. A network module detection method, modularity optimization (Guimerà and Nunes Amaral, 2005), was used to find densely connected modules in these networks.
Comparison of Time-Course Profiles
EDGE software (Storey et al., 2005) implements a significance method for analyzing time-course microarray data. To derive temporal information, we used this method to compare two time-course profiles of a given metabolite, and thereby identified all of the metabolites that showed significantly different time courses between ethanol and ABA treatment conditions, or between the wild type and gpa1. In this study, we chose the P-value cut-off as P < 0.05, and the false discovery rate–based measure of significance (the Q-value) as Q < 0.05 (Storey et al., 2005).
Comparison of Guard Cell and Mesophyll Cell Metabolite Profiles
To compare the time courses of metabolite response in mesophyll cell protoplasts versus guard cell protoplasts, we normalized the metabolite profiles of each cell type. Specifically, we divided the metabolite level at each time point by the level at 0 min, then imposed a log2 transformation on the ratio to make the ratio ranges corresponding to upregulation and downregulation (compared with 0 min) symmetric, namely [0, +∞] and [-∞, 0]. We then used EDGE software to compare the log ratio profiles of the hormone metabolite data in the two cell types. This normalization allowed us to compare the relative temporal changes of metabolite levels between the two cell types.
Analysis of Individual Time Points
The Student’s t test was performed on the log2-transformed abundances to compare metabolite levels at individual time points of treatment with ABA with the same time points under the solvent control (ethanol treatment) condition. To quantitatively represent the ABA response of metabolites in different genotypes (Col and gpa1), we used vector diagrams (Breitling et al., 2005) to show the log2-transformed P-values from the Student’s t test in two-dimensional coordinate systems (see Supplemental Figure 3 online). The coordinate systems are divided into different regions to show different ABA response patterns of metabolites.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative and GenBank/EMBL libraries under the following accession number: GPA1 (At2G26300, M32887.1).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article.
Supplemental Figure 1. Purity of Guard Cell Protoplast Preparations.
Supplemental Figure 2. Viability and ABA Responsiveness of Guard Cell Protoplasts.
Supplemental Figure 3. Distribution of Metabolites Responsive to ABA in Col and gpa1 Guard Cells.
Supplemental Data Set 1. Optimization of Metabolite Standards by MRM and Metabolite Contents (pmol mg−1 protein) of Arabidopsis Protoplasts; AB SCIEX 4000 QTRAP Calibration Notes.
Supplemental Data Set 2. PCA Analysis Loading Plots for the Data of Figure 2.
Supplemental Data Set 3. Total Guard Cell Protoplast (GCP) and Mesophyll Cell Protoplast (MCP) Numbers and Protein Contents.
Supplemental Data Set 4. Quality of the Replicates by Pair-Wise PCCs.
Supplemental Data Set 5. EDGE Analysis Confirms Similarity of gpa1-3 and gpa1-4 Guard Cell Time Courses.
Acknowledgments
This research was supported by grants from the National Science Foundation (NSF-MCB-0817954 and NSF-MCB-1167921 to S.M.A. and NSF-MCB-0818051 and NSF-MCB-1158000 to S.C.). The authors thank Ning Zhu, University of Florida, for technical assistance and advice. They also thank Ken-ichiro Hayashi, Okayama University of Science, for the kind gift of PEO-IAA.
AUTHOR CONTRIBUTIONS
S.M.A. and S.C. conceived the project. S.M.A. and X.J. designed the experiments. X.J., M.Z., and B.W.J. performed the experiments with advice from S.C. and S.M.A. R.-S.W., X.J., and R.A. performed the data analyses. S.M.A., X.J., and R.-S.W. prepared the article, with input from all authors.
References
- Abdi H., Williams L.J. (2010). Principal component analysis. Wiley Interdisc. Rev. Comput. Stat. 2: 433–459 [Google Scholar]
- Acharya B.R., Assmann S.M. (2009). Hormone interactions in stomatal function. Plant Mol. Biol. 69: 451–462 [DOI] [PubMed] [Google Scholar]
- Acharya B.R., Jeon B.W., Zhang W., Assmann S.M. (2013). Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 200: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Allen D.K., Libourel I.G.L., Shachar-Hill Y. (2009). Metabolic flux analysis in plants: Coping with complexity. Plant Cell Environ. 32: 1241–1257 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Muir S.R., Sanders D. (1995). Release of Ca2+ from individual plant vacuoles by both InsP3 and cyclic ADP-ribose. Science 268: 735–737 [DOI] [PubMed] [Google Scholar]
- Allwood J.W., Goodacre R. (2010). An introduction to liquid chromatography-mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem. Anal. 21: 33–47 [DOI] [PubMed] [Google Scholar]
- Arbona V., Argamasilla R., Gómez-Cadenas A. (2010). Common and divergent physiological, hormonal and metabolic responses of Arabidopsis thaliana and Thellungiella halophila to water and salt stress. J. Plant Physiol. 167: 1342–1350 [DOI] [PubMed] [Google Scholar]
- Assmann S.M. (December 21, 2004). Plant G proteins, phytohormones, and plasticity: three questions and a speculation. 264: re20. [DOI] [PubMed] [Google Scholar]
- Assmann S.M. (2005). G proteins Go green: A plant G protein signaling FAQ sheet. Science 310: 71–73 [DOI] [PubMed] [Google Scholar]
- Assmann S.M. (2010). Hope for Humpty Dumpty: Systems biology of cellular signaling. Plant Physiol. 152: 470–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann S.M., Grantz D.A. (1990). Stomatal response to humidity in sugarcane and soybean: Effect of vapour pressure difference on the kinetics of the blue light response. Plant Cell Environ. 13: 163–169 [Google Scholar]
- Assmann S.M., Snyder J.A., Lee Y.R.J. (2000). ABA-deficient (aba1) and ABA-insensitive (abi1–1, abi2–1) mutants of Arabidopsis have a wild-type stomatal response to humidity. Plant Cell Environ. 23: 387–395 [Google Scholar]
- Berridge M.J. (1993). Inositol trisphosphate and calcium signalling. Nature 361: 315–325 [DOI] [PubMed] [Google Scholar]
- Blatt M.R., Thiel G., Trentham D.R. (1990). Reversible inactivation of K+ channels of Vicia stomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature 346: 766–769 [DOI] [PubMed] [Google Scholar]
- Böttcher C., von Roepenack-Lahaye E., Schmidt J., Schmotz C., Neumann S., Scheel D., Clemens S. (2008). Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol. 147: 2107–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A. (2005). Vector analysis as a fast and easy method to compare gene expression responses between different experimental backgrounds. BMC Bioinformatics 6: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Gallie D.R. (2004). The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 16: 1143–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol S., Fan L.-M., Le Stunff H., Spiegel S., Gilroy S., Assmann S.M. (2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Coursol S., Le Stunff H., Lynch D.V., Gilroy S., Assmann S.M., Spiegel S. (2005). Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol. 137: 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousson A. (2003). Pharmacological evidence for a positive influence of the cyclic GMP-independent transduction on the cyclic GMP-mediated Ca2+-dependent pathway within Arabidopsis stomatal opening in response to auxin. Plant Sci. 164: 759–767 [DOI] [PubMed] [Google Scholar]
- Cousson A., Vavasseur A. (1998). Putative involvement of cytosolic Ca2+ and GTP-binding proteins in cyclic-GMP-mediated induction of stomatal opening by auxin in Commelina communis L. Planta 206: 308–314 [Google Scholar]
- Creelman R.A., Mullet J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (2011). Plant Hormones: Biosynthesis, Signal Transduction, Action! (Dordrecht: Springer). [Google Scholar]
- DellaPenna D., Last R.L. (2008). Genome-enabled approaches shed new light on plant metabolism. Science 320: 479–481 [DOI] [PubMed] [Google Scholar]
- Dubovskaya L.V., Bakakina Y.S., Kolesneva E.V., Sodel D.L., McAinsh M.R., Hetherington A.M., Volotovski I.D. (2011). cGMP-dependent ABA-induced stomatal closure in the ABA-insensitive Arabidopsis mutant abi1-1. New Phytol. 191: 57–69 [DOI] [PubMed] [Google Scholar]
- Ebert B., Zöller D., Erban A., Fehrle I., Hartmann J., Niehl A., Kopka J., Fisahn J. (2010). Metabolic profiling of Arabidopsis thaliana epidermal cells. J. Exp. Bot. 61: 1321–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers D., Lefèvre I., Legay S., Lamoureux D., Hausman J.F., Rosales R.O., Marca L.R., Hoffmann L., Bonierbale M., Schafleitner R. (2010). Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. J. Exp. Bot. 61: 2327–2343 [DOI] [PubMed] [Google Scholar]
- Fan L.M., Zhang W., Chen J.G., Taylor J.P., Jones A.M., Assmann S.M. (2008). Abscisic acid regulation of guard-cell K+ and anion channels in Gβ- and RGS-deficient Arabidopsis lines. Proc. Natl. Acad. Sci. USA 105: 8476–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2011). Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata C., Gay R., Sokolovski S., Hills A., Lamattina L., Blatt M.R. (2003). Nitric oxide regulates K+ and Cl- channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc. Natl. Acad. Sci. USA 100: 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring C.A., Irving H.R., McConchie R., Parish R.W. (1997). Jasmonates induce intracellular alkalinization and closure of Paphiopedilum guard cells. Ann. Bot. 80: 485–489 [Google Scholar]
- Gilroy S., Read N.D., Trewavas A.J. (1990). Elevation of cytoplasmic calcium by caged calcium or caged inositol triphosphate initiates stomatal closure. Nature 346: 769–771 [DOI] [PubMed] [Google Scholar]
- Goodacre R., Vaidyanathan S., Dunn W.B., Harrigan G.G., Kell D.B. (2004). Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 22: 245–252 [DOI] [PubMed] [Google Scholar]
- Guimerà R., Nunes Amaral L.A. (2005). Functional cartography of complex metabolic networks. Nature 433: 895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R. (2012). Ion channels in plants. Physiol. Rev. 92: 1777–1811 [DOI] [PubMed] [Google Scholar]
- Herde O., Peña-Cortés H., Willmitzer L., Fisahn J. (1997). Stomatal responses to jasmonic acid, linolenic acid and abscisic acid in wild-type and ABA-deficient tomato plants. Plant Cell Environ. 20: 136–141 [Google Scholar]
- Hetherington A.M., Brownlee C. (2004). The generation of Ca2+ signals in plants. Annu. Rev. Plant Biol. 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62: 2465–2483 [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., Yano M., Goodenowe D.B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101: 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefgen R., Nikiforova V.J. (2008). Metabolomics integrated with transcriptomics: Assessing systems response to sulfur-deficiency stress. Physiol. Plant. 132: 190–198 [DOI] [PubMed] [Google Scholar]
- Hossain M.A., Munemasa S., Uraji M., Nakamura Y., Mori I.C., Murata Y. (2011). Involvement of endogenous abscisic acid in methyl jasmonate-induced stomatal closure in Arabidopsis. Plant Physiol. 156: 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Mills L.N., Pical C., Leckie C.P., Aitken F.L., Kopka J., Mueller-Roeber B., McAinsh M.R., Hetherington A.M., Gray J.E. (2003). Phospholipase C is required for the control of stomatal aperture by ABA. Plant J. 34: 47–55 [DOI] [PubMed] [Google Scholar]
- Ishida T., Adachi S., Yoshimura M., Shimizu K., Umeda M., Sugimoto K. (2010). Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71 [DOI] [PubMed] [Google Scholar]
- Israelsson M., Siegel R.S., Young J., Hashimoto M., Iba K., Schroeder J.I. (2006). Guard cell ABA and CO2 signaling network updates and Ca2+ sensor priming hypothesis. Curr. Opin. Plant Biol. 9: 654–663 [DOI] [PubMed] [Google Scholar]
- Jacob T., Ritchie S., Assmann S.M., Gilroy S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 96: 12192–12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Ecker J.R., Chen J.-G. (2003). A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 131: 1623–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J.H., Wang S., Chen J.G., Jones A.M., Fedoroff N.V. (2005). Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joudoi T., Shichiri Y., Kamizono N., Akaike T., Sawa T., Yoshitake J., Yamada N., Iwai S. (2013). Nitrated cyclic GMP modulates guard cell signaling in Arabidopsis. Plant Cell 25: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.Y., Kim Y.W., Kwak J.M., Hwang J.U., Young J., Schroeder J.I., Hwang I., Lee Y. (2002). Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14: 2399–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempa S., Krasensky J., Dal Santo S., Kopka J., Jonak C. (2008). A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS One 3: e3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.K., Bamba T., Harada K., Fukusaki E., Kobayashi A. (2007). Time-course metabolic profiling in Arabidopsis thaliana cell cultures after salt stress treatment. J. Exp. Bot. 58: 415–424 [DOI] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M., et al. (2011). Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 67: 354–369 [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.-M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D.G., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L., García-Mata C., Graziano M., Pagnussat G. (2003). Nitric oxide: The versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54: 109–136 [DOI] [PubMed] [Google Scholar]
- Last R.L., Jones A.D., Shachar-Hill Y. (2007). Towards the plant metabolome and beyond. Nat. Rev. Mol. Cell Biol. 8: 167–174 [DOI] [PubMed] [Google Scholar]
- Leckie C.P., McAinsh M.R., Allen G.J., Sanders D., Hetherington A.M. (1998). Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Choi Y.B., Suh S., Lee J., Assmann S.M., Joe C.O., Kelleher J.F., Crain R.C. (1996). Abscisic acid-induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol. 110: 987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kim Y.-W., Jeon B.W., Park K.-Y., Suh S.J., Seo J., Kwak J.M., Martinoia E., Hwang I., Lee Y. (2007). Phosphatidylinositol 4,5-bisphosphate is important for stomatal opening. Plant J. 52: 803–816 [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie E.A.C., Brearley C.A. (2000). Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. USA 97: 8687–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie E.A.C., Webb A.A.R., Manison N.F., Brownlee C., Skepper J.N., Chen J., Prestwich G.D., Brearley C.A. (2003). Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc. Natl. Acad. Sci. USA 100: 10091–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Assmann S.M., Albert R. (2006). Predicting essential components of signal transduction networks: A dynamic model of guard cell abscisic acid signaling. PLoS Biol. 4: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse G., Hedrich R. (1992). Characterization of the plasma-membrane H+-ATPase from Vicia faba guard cells: Modulation by extracellular factors and seasonal changes. Planta 188: 206–214 [DOI] [PubMed] [Google Scholar]
- Lorenzo Tejedor M., Mizuno H., Tsuyama N., Harada T., Masujima T. (2012). In situ molecular analysis of plant tissues by live single-cell mass spectrometry. Anal. Chem. 84: 5221–5228 [DOI] [PubMed] [Google Scholar]
- Lynch D.V., Chen M., Cahoon E.B. (2009). Lipid signaling in Arabidopsis: No sphingosine? No problem! Trends Plant Sci. 14: 463–466 [DOI] [PubMed] [Google Scholar]
- MacRobbie E.A.C. (1998). Signal transduction and ion channels in guard cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353: 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie E.A.C. (2000). ABA activates multiple Ca2+ fluxes in stomatal guard cells, triggering vacuolar K+ Rb+ release. Proc. Natl. Acad. Sci. USA 97: 12361–12368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt F., Kemper A., Tallman G. (2001). Inhibitors of ethylene synthesis inhibit auxin-induced stomatal opening in epidermis detached from leaves of Vicia faba L. Plant Cell Physiol. 42: 223–230 [DOI] [PubMed] [Google Scholar]
- Mills L.N., Hunt L., Leckie C.P., Aitken F.L., Wentworth M., McAinsh M.R., Gray J.E., Hetherington A.M. (2004). The effects of manipulating phospholipase C on guard cell ABA-signalling. J. Exp. Bot. 55: 199–204 [DOI] [PubMed] [Google Scholar]
- Munemasa S., Mori I.C., Murata Y. (2011). Methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid in guard cells. Plant Signal. Behav. 6: 939–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S., Oda K., Watanabe-Sugimoto M., Nakamura Y., Shimoishi Y., Murata Y. (2007). The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol. 143: 1398–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navazio L., Mariani P., Sanders D. (2001). Mobilization of Ca2+ by cyclic ADP-ribose from the endoplasmic reticulum of cauliflower florets. Plant Physiol. 125: 2129–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S.J., Desikan R., Clarke A., Hancock J.T. (2002). Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol. 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Ng C.K.Y., Carr K., McAinsh M.R., Powell B., Hetherington A.M. (2001). Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410: 596–599 [DOI] [PubMed] [Google Scholar]
- Nilson S.E., Assmann S.M. (2010). The α-subunit of the Arabidopsis heterotrimeric G protein, GPA1, is a regulator of transpiration efficiency. Plant Physiol. 152: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Nakano H., Hayashi K.-i., Niwa C., Koshiba T. (2009). Differential downward stream of auxin synthesized at the tip has a key role in gravitropic curvature via TIR1/AFBs-mediated auxin signaling pathways. Plant Cell Physiol. 50: 1874–1885 [DOI] [PubMed] [Google Scholar]
- Nishiyama R., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver M.J., Guo L., Alexander D.C., Ryals J.A., Wone B.W.M., Cushman J.C. (2011). A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell 23: 1231–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X.Q., Welti R., Wang X.M. (2008). Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 1773–1781 [DOI] [PubMed] [Google Scholar]
- Pandey S., Chen J.G., Jones A.M., Assmann S.M. (2006). G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 141: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandey S., Wang R.S., Wilson L., Li S., Zhao Z., Gookin T.E., Assmann S.M., Albert R. (2010). Boolean modeling of transcriptome data reveals novel modes of heterotrimeric G-protein action. Mol. Syst. Biol. 6: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S., Wang X.Q., Coursol S.A., Assmann S.M. (2002). Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol. 153: 517–526 [DOI] [PubMed] [Google Scholar]
- Perera I.Y., Hung C.Y., Moore C.D., Stevenson-Paulik J., Boss W.F. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20: 2876–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus-Barbeoch L., Jones A.M., Assmann S.M. (2004). Plant heterotrimeric G protein function: Insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 7: 719–731 [DOI] [PubMed] [Google Scholar]
- Rao S.R., Ford K.L., Cassin A.M., Roessner U., Patterson J.H., Bacic A. (2010). Proteomic and metabolic profiling of rice suspension culture cells as a model to study abscisic acid signaling response pathways in plants. J. Proteome Res. 9: 6623–6634 [DOI] [PubMed] [Google Scholar]
- Rizhsky L., Liang H., Shuman J., Shulaev V., Davletova S., Mittler R. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema M.R.G., Hedrich R. (2010). Making sense out of Ca2+ signals: Their role in regulating stomatal movements. Plant Cell Environ. 33: 305–321 [DOI] [PubMed] [Google Scholar]
- Rubakhin S.S., Lanni E.J., Sweedler J.V. (2013). Progress toward single cell metabolomics. Curr. Opin. Biotechnol. 24: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, H., and Agell, M.I. (2000). The Analysis of Variance. (Boston: Birkhauser). [Google Scholar]
- Schilmiller A., Shi F., Kim J., Charbonneau A.L., Holmes D., Daniel Jones A., Last R.L. (2010). Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 62: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X.P., Song X.G. (2006). Cytokinin- and auxin-induced stomatal opening is related to the change of nitric oxide levels in guard cells in broad bean. Physiol. Plant. 128: 569–579 [Google Scholar]
- Shimazaki K., Doi M., Assmann S.M., Kinoshita T. (2007). Light regulation of stomatal movement. Annu. Rev. Plant Biol. 58: 219–247 [DOI] [PubMed] [Google Scholar]
- Shrestha B., Patt J.M., Vertes A. (2011). In situ cell-by-cell imaging and analysis of small cell populations by mass spectrometry. Anal. Chem. 83: 2947–2955 [DOI] [PubMed] [Google Scholar]
- Shulaev V., Cortes D., Miller G., Mittler R. (2008). Metabolomics for plant stress response. Physiol. Plant. 132: 199–208 [DOI] [PubMed] [Google Scholar]
- Snaith P.J., Mansfield T.A. (1982). Stomatal sensitivity to abscisic acid: Can it be defined? Plant Cell Environ. 5: 309–311 [Google Scholar]
- Song X., She X., He J., Huang C., Song T. (2006). Cytokinin- and auxin-induced stomatal opening involves a decrease in levels of hydrogen peroxide in guard cells of Vicia faba. Funct. Plant Biol. 33: 573–583 [DOI] [PubMed] [Google Scholar]
- Staxén I., Pical C., Montgomery L.T., Gray J.E., Hetherington A.M., McAinsh M.R. (1999). Abscisic acid induces oscillations in guard-cell cytosolic free calcium that involve phosphoinositide-specific phospholipase C. Proc. Natl. Acad. Sci. USA 96: 1779–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M., Sulpice R., Keurentjes J. (2010). Metabolic networks: How to identify key components in the regulation of metabolism and growth. Plant Physiol. 152: 428–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Xiao W., Leek J.T., Tompkins R.G., Davis R.W. (2005). Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. USA 102: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhita D., Kolla V.A., Vavasseur A., Raghavendra A.S. (2003). Different signaling pathways involved during the suppression of stomatal opening by methyl jasmonate or abscisic acid. Plant Sci. 164: 481–488 [Google Scholar]
- Suhita D., Raghavendra A.S., Kwak J.M., Vavasseur A. (2004). Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol. 134: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Hayashi K.-i., Kinoshita T. (2012). Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sano T., Tamaoki M., Nakajima N., Kondo N., Hasezawa S. (2006). Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. J. Exp. Bot. 57: 2259–2266 [DOI] [PubMed] [Google Scholar]
- Toubiana D., Fernie A.R., Nikoloski Z., Fait A. (2013). Network analysis: Tackling complex data to study plant metabolism. Trends Biotechnol. 31: 29–36 [DOI] [PubMed] [Google Scholar]
- Tsugama D., Liu S., Takano T. (2013). Arabidopsis heterotrimeric G protein β subunit, AGB1, regulates brassinosteroid signalling independently of BZR1. J. Exp. Bot. 64: 3213–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Chen J.G., Botella J.R., Jones A.M. (2013). Heterotrimeric G protein signalling in the plant kingdom. Open Biol.3: 120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K., Maruyama K., Ogata Y., Morishita Y., Takeda M., Sakurai N., Suzuki H., Saito K., Shibata D., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2009). Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 57: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Wang P., Song C.P. (2008). Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 178: 703–718 [DOI] [PubMed] [Google Scholar]
- Wang R.S., Pandey S., Li S., Gookin T.E., Zhao Z., Albert R., Assmann S.M. (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Q., Ullah H., Jones A.M., Assmann S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Worrall D., Liang Y.K., Alvarez S., Holroyd G.H., Spiegel S., Panagopulos M., Gray J.E., Hetherington A.M. (2008). Involvement of sphingosine kinase in plant cell signalling. Plant J. 56: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.J., Jin Y.J., Xu X.Y., Lu R.C., Chen H.J. (2008). Study on the extraction, purification and quantification of jasmonic acid, abscisic acid and indole-3-acetic acid in plants. Phytochem. Anal. 19: 560–567 [DOI] [PubMed] [Google Scholar]
- Zhang W., Jeon B.W., Assmann S.M. (2011). Heterotrimeric G-protein regulation of ROS signalling and calcium currents in Arabidopsis guard cells. J. Exp. Bot. 62: 2371–2379 [DOI] [PubMed] [Google Scholar]
- Zhao J., Davis L.C., Verpoorte R. (2005). Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 23: 283–333 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Stanley B.A., Zhang W., Assmann S.M. (2010). ABA-regulated G protein signaling in Arabidopsis guard cells: A proteomic perspective. J. Proteome Res. 9: 1637–1647 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Zhang W., Stanley B.A., Assmann S.M. (2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20: 3210–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Dai S., McClung S., Yan X., Chen S. (2009). Functional differentiation of Brassica napus guard cells and mesophyll cells revealed by comparative proteomics. Mol. Cell. Proteomics 8: 752–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Simons B., Zhu N., Oppenheimer D.G., Chen S. (2010). Analysis of abscisic acid responsive proteins in Brassica napus guard cells by multiplexed isobaric tagging. J. Proteomics 73: 790–805 [DOI] [PubMed] [Google Scholar]