CELLULOSE SYNTHASE INTERACTIVE3 (CSI3) is an important regulator of cellulose biosynthesis and anisotropic growth. Together with CSI1, CSI3 regulates the rate of cellulose biosynthesis in both a microtubule-dependent and microtubule-independent manner in Arabidopsis.
Abstract
Anisotropic plant cell growth depends on the coordination between the orientation of cortical microtubules and the orientation of nascent cellulose microfibrils. CELLULOSE SYNTHASE INTERACTIVE1 (CSI1) is a key scaffold protein that guides primary cellulose synthase complexes (CSCs) along cortical microtubules during cellulose biosynthesis. Here, we investigated the function of the CSI1-like protein, CSI3, in Arabidopsis thaliana. Similar to CSI1, CSI3 associates with primary CSCs in vitro, colocalizes with CSCs in vivo, and exhibits the same plasma membrane localization and bidirectional motility as CSI1. However, ProCSI1:GFP-CSI3 cannot complement the anisotropic cell growth defect in csi1 mutants, suggesting that CSI3 is not functionally equivalent to CSI1. Also, the colocalization ratio between CSI1 and CSI3 is low, which may suggest heterogeneity within the CSC population. csi1 csi3 double mutants showed an enhanced cell expansion defect as well as an additive reduction of CSC velocities, and CSI3 dynamics are dependent on CSI1 function. We propose that CSI3 is an important regulator of plant cellulose biosynthesis and plant anisotropic cell growth that modulates the velocity of CSCs in both a microtubule-dependent and microtubule-independent manner.
INTRODUCTION
A central question in plant cell development is how the cell wall, which is the equivalent of the extracellular matrix of mammalian cells, determines directional cell expansion and the final shape of the cell. Cellulose microfibrils, the major load-bearing component of the cell wall, are synthesized by large, plasma membrane–localized, sixfold symmetric, rosette protein complexes known as cellulose synthase (CESA) complexes (CSCs) (Kimura et al., 1999). Cellulose microfibrils are laid down transversely to the axis of elongation, thus forming a spring-like structure that reinforces the cell laterally and favors longitudinal expansion in most growing cells (Green, 1962). The mechanism by which plant cells establish and maintain the transverse orientation of cellulose microfibrils during cell expansion is controversial (Lloyd, 2011; Baskin and Gu, 2012). Within the cell, cortical microtubules are self-organized into an array near the inner surface of the plasma membrane that is aligned in parallel with the cellulose microfibrils of the extracellular cell wall (Ledbetter and Porter, 1963; Hepler and Newcomb, 1964; Dixit and Cyr, 2004; Chan et al., 2007; Wightman and Turner, 2007). The field is divided between proponents of the alignment hypothesis (i.e., that CSCs synthesize cellulose microfibrils under the guidance of cortical microtubules) and those who believe that cellulose microfibrils are organized by an intrinsic self-assembly mechanism after synthesis (Heath, 1974; Hepler and Palevitz, 1974; Roland et al., 1975; Neville et al., 1976).
Mounting evidence supports the alignment hypothesis (Herth, 1980; Giddings and Staehelin, 1991; Baskin, 2001; Paredez et al., 2006; Lloyd and Chan, 2008; Baskin and Gu, 2012; Bringmann et al., 2012; Li et al., 2012); however, it is uncertain whether microtubules can guide a full variety of microfibril alignment (Lloyd, 2011). A pivotal step forward in the analysis of cellulose deposition was the implementation of spinning disc confocal microscopy to analyze the dynamics of fluorescent protein tagged CESAs in living cells (Paredez et al., 2006). With this advancement, CSC movement and trajectories can be observed directly, and the organization of nascent cellulose microfibrils can be studied while the microfibrils are being synthesized instead of deducing microfibril orientation from micrographs of fixed specimens (Ledbetter and Porter, 1963; Hepler and Newcomb, 1964; Neville et al., 1976) or by polarized-light microscopy (Baskin et al., 2004). Studies of yellow fluorescent protein (YFP)-CESA6 dynamics have largely supported the alignment hypothesis by showing that plasma membrane–localized YFP-CESA6 particles travel along tracks that were coincident with cortical microtubules (Paredez et al., 2006). Furthermore, when seedlings were irradiated with blue light to induce the reorientation of cortical microtubules, the position and trajectories of YFP-CESA6–labeled CSCs were likewise reoriented. Although CSCs continued to travel in oblique orientations when treated with the microtubule-depolymerizing drug oryzalin, 10 to 16 h of oryzalin treatment resulted in reduced CSC motility, which suggests that microtubules may also affect the velocity of CSCs (Paredez et al., 2006; Li et al., 2012). Microtubules have also been suggested to be the target for the delivery of CSCs to the plasma membrane (Crowell et al., 2009; Gutierrez et al., 2009).
While the alignment hypothesis has garnered much support, the precise molecular mechanism by which CSCs are guided along cortical microtubules was not discovered until recently (Gu et al., 2010; Gu and Somerville, 2010; Bringmann et al., 2012; Li et al., 2012). CELLULOSE SYNTHASE INTERACTIVE1 (CSI1), initially identified through a yeast two-hybrid screen for CESA interactive proteins, interacts with both primary CESAs and microtubules to act as a key player required for the guidance of primary CSCs along microtubules during cellulose synthesis (Gu et al., 2010; Baskin and Gu, 2012; Lei et al., 2012a; Li and Gu, 2012; Li et al., 2012). However, the direct association between CSC and CSI1 has not been demonstrated in vivo. Arabidopsis thaliana encodes two CSI1-like proteins, namely, CSI2 and CSI3. Here, we investigated the roles of CSI1-like proteins in the regulation of cellulose biosynthesis, with an emphasis on the coalignment between CSCs and microtubules and the microtubule-dependent regulation of CSC velocity.
RESULTS
CSI3 and CSI1 Share Similar Expression Patterns
The Arabidopsis genome contains two CSI1 homologs, referred to as CSI2 and CSI3 (Gu et al., 2010). CSI1 shares 61 and 66% amino acid similarity with CSI2 and CSI3, respectively. CSI2 and CSI3 each have at least six predicted armadillo repeats and a single C2 domain (see Supplemental Figure 1A online). Transcriptional analysis obtained from Genevestigator indicates that CSI3, like CSI1, is widely expressed in many tissues, whereas CSI2 expression is limited to male reproductive tissues, including the stamen and pollen (see Supplemental Figure 1B online). To obtain detailed expression profiles of CSI2 and CSI3, a 3-kb promoter of CSI2 or CSI3 was fused to the β-glucuronidase (GUS) gene, and a histochemical analysis of GUS activity was performed. ProCSI3:GUS expression was detected in many of the same tissues as CSI1, including dark-grown hypocotyls, stamen, pollen, developing siliques, and roots (see Supplemental Figure 2 online). However, the expression of CSI3 was not identical to that of CSI1. In rosette leaves, CSI3 was expressed in the vasculature and in the trichomes, whereas CSI1 was not expressed in these tissues (Gu et al., 2010). In addition, the expression of CSI3 was more restricted in meristematic tissue of the shoot and root. GUS activity for ProCSI2:GUS was not detected in any of the tissues examined, including rosette leaves, 10-d-old light-grown whole seedlings, 4-d-old dark-grown seedlings, flowers, siliques, stamen, and pollen. This evidence suggests that CSI2 is a pseudogene or that the expression of CSI2 is below the detection limit of the histochemical assay.
csi1 csi3 Double Mutants Show Enhanced Cell Expansion Defects
To investigate the biological function of CSI3, we analyzed T-DNA insertion lines of CSI3. csi3-1 was determined to be a null allele, since a T-DNA was inserted in its third exon and no CSI3 mRNA was detected by RT-PCR (see Supplemental Figure 3 online). Unlike csi1 mutants, csi3-1 had no visible defects in expansion. Dark-grown hypocotyls, light-grown seedlings, and mature plants of csi3-1 were indistinguishable from the wild type.
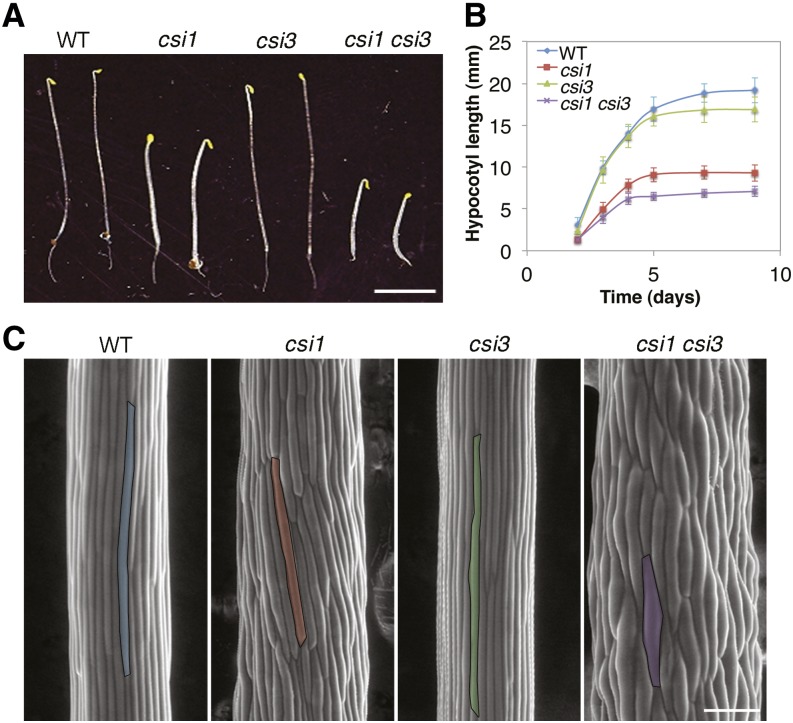
To test for genetic interaction between CSI1 and CSI3, a csi1 csi3 double mutant line was generated by crossing csi1-3 and csi3-1. csi1 csi3 etiolated hypocotyls displayed severe cell swelling and growth retardation compared with the single mutant parental lines (Figure 1). Dark-grown hypocotyls of csi1csi3 were ∼30% shorter than csi1 (Figures 1A and 1B), indicating that CSI1 and CSI3 synergistically regulate hypocotyl elongation. The number of epidermal cells in hypocotyls of csi1 csi3 did not differ significantly from that of the csi1 mutant. More importantly, csi1 csi3 exacerbated the anisotropic growth defect of csi1 hypocotyls, as illustrated by an ∼15% increase in cell diameter (see Supplemental Figure 4A online; Figure 1C). The reduced hypocotyl length and increased hypocotyl diameter of csi1 csi3 mutants indicate that CSI3 is a regulator of anisotropic growth.
Figure 1.
Morphology of csi Mutants.
(A) Four-day-old dark-grown seedlings of wild type (WT; Columbia-0), csi1, csi3, and csi1 csi3. Bar = 5 mm.
(B) Quantification of hypocotyl length of dark-grown Columbia-0, csi1, csi3, and csi1 csi3. Data were collected from the measurement of ∼50 seedlings for each genotype. Error bars represent standard error.
(C) Scanning electron micrograph of 4-d-old dark-grown hypocotyls in the wild type (Columbia-0), csi1, csi3, and csi1 csi3. One epidermal cell of each genotype is highlighted in color. Bar =100 μm.
[See online article for color version of this figure.]
Both CSI1 and CSI3 Are Required for Normal Velocity of CSCs
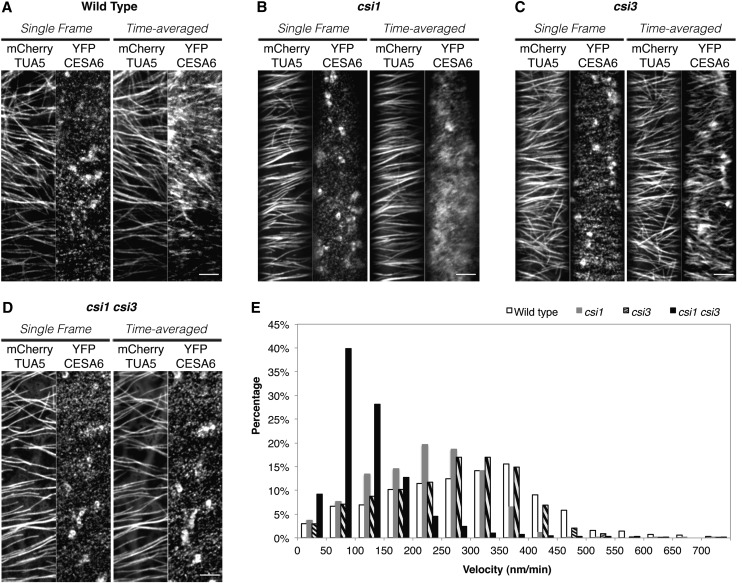
To investigate whether CSI3 has a role in the guidance of CSCs along cortical microtubules, we compared the association of CSCs with microtubules in wild-type, csi1, csi3, and csi1 csi3 lines. Using a transgenic line coexpressing the CSC marker, YFP-CESA6, and the microtubule marker mCherry-TUBULIN5 (TUA5), CSCs were visualized moving through the plasma membrane along linear tracks provided by the underlying microtubules in wild-type, dark-grown hypocotyls (Paredez et al., 2006) (Figure 2A). Quantification of the association between CSCs and microtubules revealed that YFP-CESA6 colocalized with microtubules extensively in the wild-type line (Li et al., 2012) (Table 1). The coalignment of YFP-CESA6 and microtubules was disrupted in the csi1 mutants (Li et al., 2012) (Figure 2B). Unlike csi1, csi3-1 did not affect the colocalization ratio of CSCs with microtubules (Figures 2A and 2C, Table 1). Moreover, the colocalization ratio of CSCs with microtubules in csi1 csi3 was not significantly different from that of csi1 (Figures 2B and 2D, Table 1), suggesting that unlike CSI1, CSI3 is not crucial for the coalignment of CSCs and microtubules.
Figure 2.
Both CSI1 and CSI3 Influence the Velocity of CSCs.
(A) to (D) Arabidopsis seedlings expressing both YFP-CESA6 and mCherry-TUA5 in control seedlings. (A), csi1 (B), csi3 (C), and csi1 csi3 (D) were grown in darkness for 3 d before imaging. Epidermal cells ∼2 mm below the apical hook were imaged by confocal microscopy. A single optical section was acquired at the plane of the plasma membrane. The time-averaged images were projections of 61 frames (2-min duration; 2-s interval). Shown are representative images from ∼25 cells from 10 seedlings for each genotype. Bars = 5 μm.
(E) Histogram of particle velocities. The mean velocity is 301 ± 130 nm/min in the wild type (n = 1702), 223 ± 96 nm/min in csi1 (n = 1537), 265 ± 113 nm/min in csi3 (n = 1393), and 121 ± 80 nm/min in csi1 csi3 mutants (n = 1537).
Table 1. Quantification of Colocalization between CSCs and Microtubules in csi Mutants.
| Untreated Seedlings | The Wild Type | csi3 | csi1csi3 |
|---|---|---|---|
| No. of colocalized voxels | 353 | 307 | 222 |
| Percentage of CESA6 colocalized with microtubule | 74% ± 4% | 75% ± 3% | 35% ± 9% |
| P value | <0.001 | <0.001 | 0.606 |
| Percentage of expected random colocalized | 46% ± 3% | 47% ± 1% | 36% ± 6% |
To test whether CSI3 has a role in cellulose biosynthesis, we examined the velocity of YFP-CESA6–labeled CSCs in csi3. Similar to in the wild-type background, time-averaged projections of 61 frames from a 2-min time series in csi3 displayed linear trajectories along microtubules (Figures 2A and 2C). The velocity of CSCs was quantified by measuring the slopes of linear traces in kymographs (Paredez et al., 2006). The velocity of YFP-CESA6 in csi3 was not significantly different from that of control cells (Figure 2E). By contrast, the velocity of YFP-CESA6 in csi1 and csi1 csi3 mutants was reduced (Figures 2B and 2D). The mean velocity of YFP-CESA6 in csi1 was 223 ± 96 nm/min (n = 1537) compared with 283 ± 137 nm/min (n = 902) in control seedlings. Loss of both CSI1 and CSI3 further reduced the velocity of CSCs to 121 ± 80 nm/min in csi1csi3, a reduction of ∼45% from the average velocity in csi1 mutants, suggesting that CSI3 is required for maintaining the velocity of CSCs in csi1 mutants. Crystalline cellulose analysis using the Updegraff method suggested that the cellulose content in etiolated hypocotyls of csi1 csi3 was further reduced than in csi1 (see Supplemental Figure 4B online). These observations are consistent with the enhanced anisotropic growth defects in csi1 csi3 seedlings.
CSI3 Temporarily Associates with CSI1
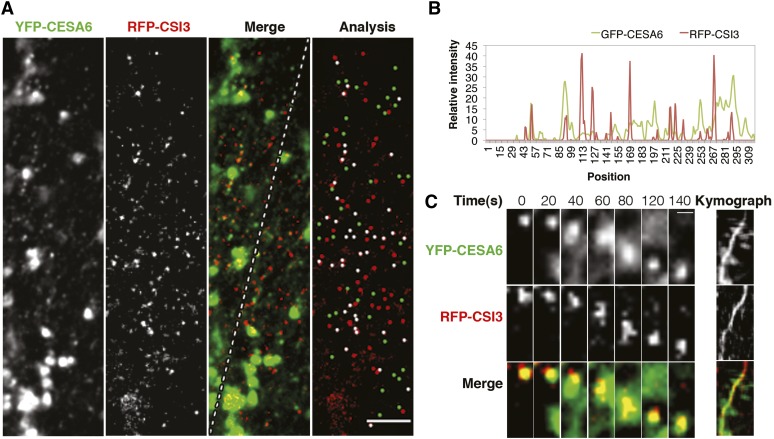
To examine the subcellular localization of CSI3, we created an N-terminal green fluorescent protein (GFP) fusion construct using full-length cDNA of CSI3 driven by a 3-kb native promoter of CSI3 (see Supplemental Figure 5A online). To test the reliability of this variant, the resulting ProCSI3:GFP-CSI3 construct was transformed into the csi1 csi3 double mutant. Since csi3 lacked a measureable phenotype, complementation analysis was performed on the double mutant. Expression of GFP-CSI3 in dark-grown seedlings csi1 csi3 complemented the csi3 mutation and resulted in seedlings with a csi1 morphology (see Supplemental Figures 5B to 5D online), indicating that GFP-CSI3 fusion proteins are functional in plants. Similar to GFP-CSI1, GFP-CSI3 was detected as distinct particles at the plasma membrane of epidermal cells of dark-grown hypocotyls and was not detected at intracellular Golgi-associated compartments (see Supplemental Figure 5E online). To analyze the relationship between CSI1 and CSI3, we crossed a GFP-CSI3 line with red fluorescent protein (RFP)-CSI1 (Li et al., 2012) and observed epidermal cells of 3-d-old dark-grown hypocotyls via two-channel confocal imaging. RFP-CSI1 and GFP-CSI3 particles moved along linear tracks, as shown by time-averaged projections of 61 frames from a 5-min time series of images (Figure 3A; see Supplemental Movie 1 online). The linear tracks traveled by GFP-CSI3 coincided with those of RFP-CSI1, as shown by the merged image (Figure 3A). Both GFP-CSI1 and GFP-CSI3 particles traveled bidirectionally as shown by cross-hatching traces in the kymographs (Figure 3B). The average velocity of GFP-CSI3 and RFP-CSI1 particles was similar to that of GFP-CSI3 particles traveling at 374 ± 141 nm/min (range 50 to 550, n = 554) and RFP-CSI1 traveling at 377 ± 114 nm/min (range 50 to 600, n = 633) in the same cells (Figure 3C).
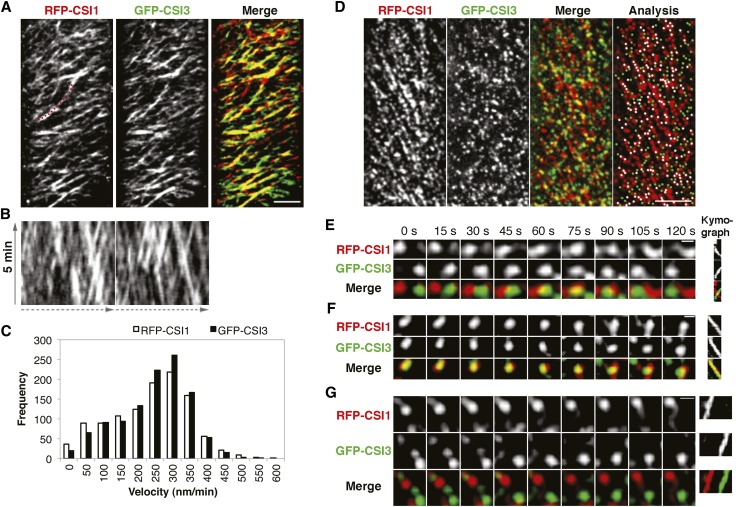
Figure 3.
CSI3 Partially Colocalizes with CSI1.
Arabidopsis seedlings expressing both RFP-CSI1 and GFP-CSI3 were grown in dark for 3 d before imaging. Epidermal cells ∼2 mm below the apical hook were imaged by confocal microscopy.
(A) Time average of 61 frames (5-min duration; 5-s interval) showing linear trajectories of RFP-CSI1, GFP-CSI3, and merge image. A representative image from 45 cells from 15 seedlings is shown. Bar = 5 μm.
(B) Kymographs of a linear path highlighted in (A), displaying steady and bidirectional movement for both RFP-CSI1 (left) and GFP-CSI3 (right).
(C) Histogram of particle velocities. The mean velocity is 374 ± 141 nm/min for RFP-CSI1 (n = 544) and 377 ± 114 nm/min for GFP-CSI3 (n = 633).
(D) A single section image of RFP-CSI1 and GFP-CSI3 and colocalization analysis of the merged image. Shown is a representative image from 45 cells from 15 seedlings. White dots (n = 176) represent colocalized RFP-CSI1 and GFP-CSI3. RFP-CSI1 or GFP-CSI3 that did not colocalize is colored in red (n = 174) and green (n = 181), respectively. Bar = 5 μm.
(E) and (F) RFP-CSI1 and GFP-CSI3 particles show dynamic behavior at the plasma membrane.
(E) At the start (t = 0 s), RFP-CSI1 and GFP-CSI3 particles were in opposite corners and moved toward one another. At 75 s, RFP-CSI1 and GFP-CSI3 particles overlapped with one another and then moved in opposite directions and reached the opposite corners at 120 s. Shown is one representative instance chosen from 11 documented events. Bar = 0.5 μm.
(F) From the start (t = 0 s) to the end (t = 120 s), RFP-CSI1 and GFP-CSI3 particles were colocalized and moved in the same direction. One representative instance chosen from nine documented events is shown. Bar = 0.5 μm.
(G) RFP-CSI1 and GFP-CSI3 particles moved in the same direction but one (GFP-CSI3 in this case) lagged behind the other. One representative instance chosen from >35 documented events is shown. Bar = 0.5 μm.
[See online article for color version of this figure.]
RFP-CSI1 and GFP-CSI3 particles partially overlapped at the plasma membrane (Figure 3D). In a single optical section, 43% ± 3% of GFP-CSI3 particles (six cells from six individual seedlings; Table 2) were colocalized with RFP-CSI1, which was not significantly higher than the random colocalization (38% ± 2%, P value of 0.074). Similarly, 52% ± 3% of RFP-CSI1 particles (six cells from six individual seedlings; Table 2) were colocalized with GFP-CSI3, which was just slightly higher than the random colocalization (39% ± 2%, P value 0.003). These results suggest that the colocalization of RFP-CSI1 and GFP-CSI3 is not widespread at the plasma membrane.
Table 2. Quantification of Colocalization among CSI1, CSI3, CSCs, and Microtubules.
| Untreated Seedlings | RFP-CSI1 (A) vs. | YFP-CESA6 (A) vs. | GFP-CSI3 (A) vs. |
|---|---|---|---|
| GFP-CSI3 (B) | CSI3-RFP (B) | RFP-TUA5 (B) | |
| No. of colocalized voxels | 362 | 470 | 685 |
| Percentage of materia A/B colocalized | 52% ± 3%/43% ± 3% | 75% ± 3%/75% ± 3% | 80% ± 3% |
| P value | 0.003/0.074 | <0.001 | <0.001 |
| Percentage of expected random colocalized | 39% ± 2%/38% ± 2% | 33% ± 1%/35% ± 2% | 46% ± 2% |
Given that RFP-CSI1 and GFP-CSI3 each travel bidirectionally with similar constant velocities along overlapping tracks, the limited colocalization between CSI1 and CSI3 could result either from RFP-CSI1 and GFP-CSI3 particles traveling in opposite directions or from RFP-CSI1 and GFP-CSI3 traveling in the same direction while maintaining different positions along the same track. To decipher the relationship between CSI1 and CSI3, we examined the temporal dynamic behavior of RFP-CSI1 and GFP-CSI3 particles. In 65% of observed cases (51 out of 78 colocalization events), RFP-CSI1 and GFP-CSI3 particles moved in opposite directions, overlapping briefly (at the 75-s time point) before parting and continuing to move in opposite directions (Figure 3E). Furthermore, we observed prolonged associations between RFP-CSI1 and GFP-CSI3 particles that were moving in the same direction, in which colocalized particles traveled together for at least 2 min before splitting into distinct particles (27 out of 78 colocalization events; Figure 3F; see Supplemental Movie 2 online). However, in the majority of cases in which RFP-CSI1 and GFP-CSI3 particles moved in the same direction, one particle lagged behind the other (Figure 3G; see Supplemental Movie 2 online). These observations suggest that the limited colocalization between RFP-CSI1 and GFP-CSI3 may represent particles that maintain distinct roles, while temporarily overlapping without functional association.
CSI3 Associates with the Primary CSCs
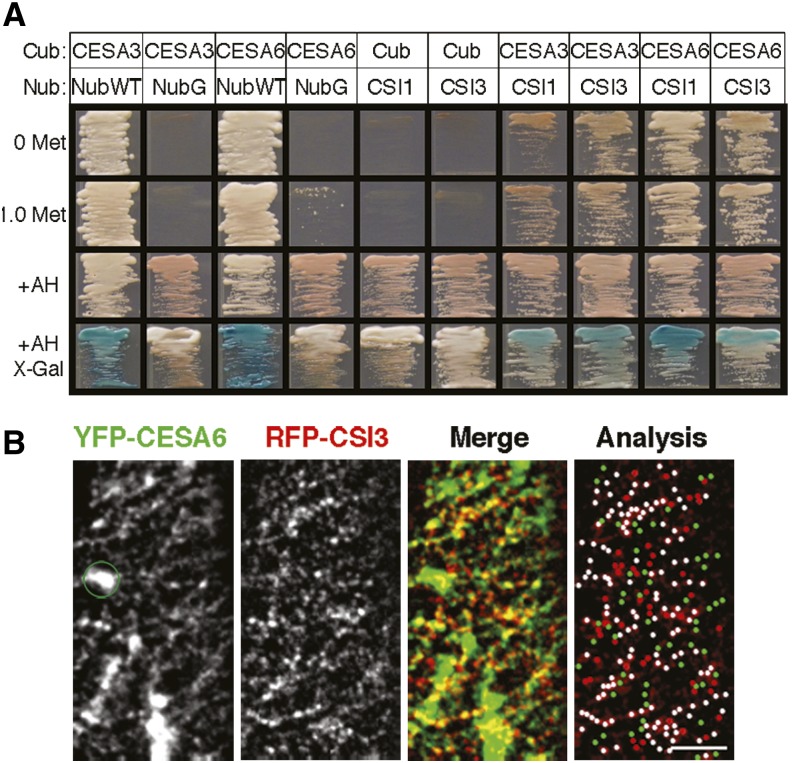
CSI1 interacts with the primary CESAs, including CESA1, CESA3, and CESA6, in a conventional yeast two-hybrid assay (Gu et al., 2010; Gu and Somerville, 2010). To test for direct interaction between CSI3 and the full-length CESAs, we performed a split-ubiquitin yeast two-hybrid assay, which can analyze protein–protein interactions of integral membrane proteins (Obrdlik et al., 2004). Full-length CESA3 and CESA6 sequences were fused to the C-terminal half of ubiquitin (Cub) and were able to reconstitute a functional ubiquitin when cotransformed with the wild-type N-terminal half of ubiquitin (NubWT), which was used as a positive control, but not when cotransformed with a mutated form of Nub (NubG), which was used as a negative control (Figure 4A). As an additional negative control, no reporter gene expression was detected when either CSI1-NubG or CSI3-NubG were cotransformed with Cub alone (Figure 4A). When either CSI1 or CSI3 was fused to NubG and cotransformed into yeast with CESA3-Cub or CESA6-Cub, reporter gene expression was detected, suggesting that CSI1 and CSI3 interact with full-length CESA3 and CESA6 in the split-ubiquitin yeast two-hybrid assay.
Figure 4.
CSI3 Associates with Primary CSCs Both in Vitro and in Vivo.
(A) CSI3 interacts with primary CESAs in a split-ubiquitin yeast two-hybrid assay. Interactions were selected on selective minimal medium with or without 1.0 mM Met. +AH shows the growth of yeast on nonselective minimal medium. X-gal staining reveals β-galactosidase activity. For Cub-CESAs, Nub-WT and NubGX33 were used as positive and negative controls, respectively. For CSI1NubG and CSI3NubG, empty Cub vectors were used as negative controls.
(B) Colocalization of CSI3 and CSCs. Arabidopsis seedlings expressing both RFP-CSI3 and YFP-CESA6 were grown in the dark for 3 d before imaging. Representative single optical sections of 56 cells from 20 RFP-CSI3 and YFP-CESA6 seedlings are shown. The green circle marks Golgi-localized YFP-CESA6 (absent in the RFP-CSI3 image) that was excluded from the colocalization analysis. In the analysis image, white dots (n = 134) represent colocalized RFP-CSI3 and YFP-CESA6. RFP-CSI3 or YFP-CESA6 that did not colocalize is in red (n = 50) and green (n = 56), respectively. Bar = 5 μm.
[See online article for color version of this figure.]
To examine whether CSI3 associates with CSCs in vivo, we generated a line carrying both RFP-CSI3 and YFP-CESA6. Two-channel confocal imaging revealed that the RFP-CSI3 signal extensively overlapped with YFP-CESA6 (Figure 4B; see Supplemental Movie 3 online). The colocalization quantification showed that ∼75% ± 3% of YFP-CESA6 particles (five cells from five individual seedlings) colocalized with RFP-CSI3 particles (Table 2), similar to what has been observed for the colocalization ratio between GFP-CESA6 and RFP-CSI1 (Lei et al., 2012b; Li et al., 2012). These results suggest that both CSI1 and CSI3 associate with the primary CSCs at the plasma membrane.
CSI3 Localizes to SmaCCs/MASCs
In addition to associating with CESA at the plasma membrane, CSI1 also labels small CESA-containing compartments (SmaCCs) or microtubule-associated cellulose synthase compartments (MASCs) (Lei et al., 2012b). After dark-grown hypocotyls were treated with 100 nM isoxaben for 2 h, RFP-CSI3 also accumulated in a subset of SmaCCs/MASCs that were colabeled with YFP-CESA6 (Figure 5A). RFP-CSI3–containing SmaCCs/MASCs moved with a variable speed from 10 to 3000 nm/min (see Supplemental Figure 6D online), which is consistent with the dynamics of SmaCCs/MASCs (Crowell et al., 2009; Gutierrez et al., 2009). RFP-CSI3 punctae moved simultaneously with SmaCCs/MASCs that were colabeled with YFP-CESA6 (Figure 5C; see Supplemental Movie 4 online). RFP-CSI3 punctae appeared in the same focal plane as YFP-CESA6 and showed typical erratic motility with rapid and short dislocations.
Figure 5.
CSI3 Associates with SmaCCs/MASCs.
Arabidopsis seedlings expressing both YFP-CESA6 and RFP-CSI3 were treated with 100 nM isoxaben for 2 h.
(A) Single optical section of YFP-CESA6, RFP-CSI3, and merged image and colocalization analysis of YFP-CESA6 and RFP-CSI3. Shown is a representative image from 26 cells from 11 seedlings. White dots (n = 45) represent colocalized RFP-CSI3 and YFP-CESA6. RFP-CSI3 or YFP-CESA6 that did not colocalize is colored in red (n = 44) and green (n = 40), respectively. Bar = 5 μm.
(B) Plot of a line scan showing the spatial correlation between RFP-CSI3 and YFP-CESA6 punctae. The line is indicated in the merged image in (A).
(C) Dynamic behavior of SmaCCs/MASCs. Note that SmaCCs/MASCs labeled with YFP-CESA6 and RFP-CSI3 displayed typical erratic motility, with rapid and short dislocations, as shown in the kymograph. Bar = 0.5 μm.
[See online article for color version of this figure.]
The colocalization ratio of SmaCCs/MASCs colabeled with YFP-CESA6 and RFP-CSI3 was smaller than that of SmaCCs/MASCs colabeled with YFP-CESA6 and RFP-CSI1 (Table 3), suggesting that CSI3 associates with a subset of SmaCCs/MASCs. To investigate whether the CSI1- and CSI3-containing SmaCC/MASC populations overlap, a colocalization analysis was performed between RFP-CSI1 and GFP-CSI3. The colocalization ratio of the isoxaben-induced SmaCCs/MASCs colabeled with RFP-CSI1 and GFP-CSI3 was similar to the colocalization ratio between plasma membrane–localized RFP-CSI1 and GFP-CSI3 particles in the absence of isoxaben (see Supplemental Figures 6A and 6B online; Table 3). To determine whether there was a functional relationship between CSI1 and CSI3 in colabeled SmaCCs/MASCs, the temporal behavior of CSI1- and CSI3-containing SmaCCs/MASCs was analyzed. Multiple distinct RFP-CSI1 and GFP-CSI3 isoxaben-induced SmaCC/MASC particles merged into a single SmaCC/MASC particle with overlapping RFP-CSI1 and GFP-CSI3 signal. This colabeled SmaCC/MASC particle was stable for ∼40 s before splitting into two separate RFP-CSI1–labeled particles that were distinct from the GFP-CSI3–labeled particle that remained stable and stationary (see Supplemental Figure 6C and Supplemental Movie 5 online). Similar to the relationship observed between overlapping plasma membrane–localized CSI1 and CSI3 particles, the temporal behavior of RFP-CSI1 and GFP-CSI3 colabeled SmaCCs/MASCs suggests that CSI1 and CSI3 mark two distinct SmaCC/MASC populations. Together, these results indicate that heterogeneity may exist within CSC populations and within SmaCC/MASC populations.
Table 3. Quantification of Colocalization between CESA, CSI1, and CSI3 upon Isoxaben Treatment.
| Isoxaben (2 h) | YFP-CESA6 (A) vs. RFP-CSI1 (B) | YFP-CESA6 (A) vs. RFP-CSI3 (B) | RFP-CSI1 (A) vs. GFP-CSI3 (B) |
|---|---|---|---|
| No. of colocalized voxels | 209 | 152 | 433 |
| Percentage of material A/B colocalized | 79% ± 6%/74% ± 9% | 56% ± 6%/57% ± 6% | 50% ± 4%/49% ± 7% |
| P value | <0.001 | <0.001 | <0.001 |
| Percentage of expected random colocalized | 15% ± 4%/18% ± 1% | 14% ± 6%/18% ± 3% | 18% ± 4%/19% ± 4% |
Colocalization Analysis of CSI1, CSI3, and Microtubules
CSI1 is the scaffold between CSCs and cortical microtubules (Li et al., 2012). Considering the similarity between CSI1 and CSI3, the spatial relationship between CSI3 and microtubules was examined. Two-channel confocal imaging of a line carrying both GFP-CSI3 and RFP-TUA5 revealed that CSI3 moved in a linear track along underlying cortical microtubules (see Supplemental Figure 7A and Supplemental Movie 6 online). In a single optical section, around 80% ± 3% of RFP-CSI3 particles (five cells from five individual seedlings) coaligned with microtubules. To further examine the spatial relationship between CSI1, CSI3, and microtubules, we generated a line carrying cyan fluorescent protein (CFP)-TUA1, RFP-CSI1, and GFP-CSI3. Three-channel confocal imaging revealed that the percentage of coincidence between RFP-CSI1 and microtubules (85% ± 10%, three cells from three individual seedlings) is similar to that of between GFP-CSI3 and microtubules (77% ± 10%, three cells from three individual seedlings; see Supplemental Figure 7B online). These observations suggest that, similar to CSI1, CSI3 particles travel along the underlying microtubules.
CSI3 Is Not Functionally Equivalent to CSI1 but Is Partially Dependent on CSI1
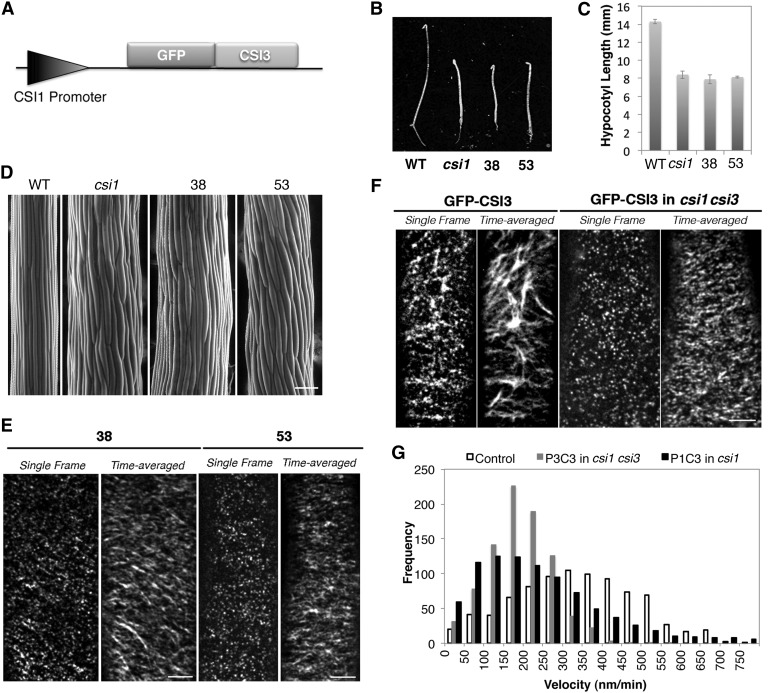
To further investigate the function of CSI3, we generated a construct in which CSI3 cDNA was fused with GFP under the control of the CSI1 promoter (ProCSI1:GFP-CSI3) (Figure 6A). The ProCSI1:GFP-CSI3 construct was transformed into csi1-3. Homozygous transgenic lines were selected and analyzed for dark-grown growth morphology. ProCSI1:GFP-CSI3 was not able to rescue the short and swollen hypocotyl phenotype of csi1-3 (Figures 6B to 6D). To assess whether ProCSI1:GFP-CSI3 (P1C3) is able to associate with CSCs at the plasma membrane, two individual lines of P1C3 in csi1-3 (#38 and #53) were examined using spinning disk confocal microscopy. P1C3 particles were localized at the plasma membrane in a pattern indistinguishable from GFP-CSI3 particles driven by the native CSI3 promoter (Figures 6E and 6F; see Supplemental Figure 5E online). P1C3 particles moved bidirectionally and formed linear trajectories, suggesting that P1C3 particles were functionally associated with CSCs. However, since P1C3 was not able to complement the mutant phenotype of csi1-3, CSI3 is not functionally equivalent to CSI1.
Figure 6.
CSI3 Is Not Functionally Equivalent to CSI1 but Is Partially Dependent on CSI1.
(A) Schematic representation of the ProCSI1:GFP-CSI3 construct.
(B) Morphology of 4-d-old, dark-grown seedlings. From left to right: the wild type (WT; Columbia), csi1, ProCSI1:GFP-CSI3 in csi1 line #38 (38), and ProCSI1:GFP-CSI3 in csi1 line #53 (53). Bar = 5 mm.
(C) Quantification of hypocotyl length of dark-grown seedlings shown in (B). Data were collected from the measurement of ∼50 seedlings for each genotype.
(D) Scanning electron micrograph of 4-d-old, dark-grown hypocotyls in the wild type (Columbia-0), csi1, and csi1 lines #38 and #53 (from left to right). Bar =100 μm.
(E) Single optical section and time average of 61 frames (5-min duration; 5-s interval) of ProCSI1:GFP-CSI3 in csi1. Bar = 5 μm.
(F) GFP-CSI3 localization and motility in control and csi1 csi3 seedlings. Single frames were acquired at the plane of the plasma membrane. Time-averaged stacks of 61 frames (5-min duration; 5-s interval) show movement of GFP-CSI3 along linear trajectories in control and csi1 csi3 seedlings. A representative image from 22 cells from nine seedlings is shown. Bar = 5 μm.
(G) Histogram of measured particle velocities in ProCSI1:GFP-CSI3 (P1C3) in csi1 and ProCSI3:GFP-CSI3 (P3C3) in csi1 csi3. The mean particle velocity is 339 ± 155 nm/min in control plants (n = 854), 194 ± 82 nm/min in csi1 csi3 (n = 864), and 261 ± 228 nm/min in csi1 (n = 874).
To test whether the function of CSI3 is dependent on CSI1, we examined the distribution and dynamics of GFP-CSI3 particles in the ProCSI3:GFP-CSI3 (P3C3) complemented csi1 csi3 mutant background. In csi1 csi3, GFP-CSI3 particles were localized at the plasma membrane and formed short and dense trajectories in the time-averaged images (Figure 6F). The average velocity of GFP-CSI3 had more than a 40% reduction when CSI1 was absent. The mean velocity of GFP-CSI3 in csi1 csi3 (194 ± 82 nm/min; Figure 6G; see Supplemental Movie 7 online) was similar to that of YFP-CESA6 in csi1 (Gu et al., 2010), suggesting that CSI1 is required for normal motility of CSI3-associated CSCs. The velocity reduction of P1C3 in csi1 csi3 is comparable to that of P3C3 in csi1 csi3 (Figure 6G), further supporting the notion that CSI1 is required for CSI3-associated CSCs.
DISCUSSION
With six putative armadillo repeat domains and a single C-terminal C2 domain, CSI3 shares 66% similarity with CSI1 in amino acid sequence. CSI1 interacted with multiple primary CESAs in a yeast two-hybrid assay (Gu et al., 2010), and CSI3 interacted with multiple primary CESAs in a split-ubiquitin yeast two-hybrid assay. The behavior of fluorescent protein fusions of CSI3 and CSI1 was similar. CSI1 and CSI3 each localized to the plasma membrane as discrete particles that colocalized with CSCs and traveled along cortical microtubule tracks at comparable velocities. However, despite these similarities, CSI3 appears to have a role that is distinct from CSI1 based on several observations. Unlike csi1 mutants, which exhibit short and swollen dark-grown hypocotyls, csi3 mutants do not exhibit an apparent phenotype. Also, in contrast with csi1 mutants, which exhibit CSCs that travel at reduced velocities along tracks that are no longer organized along microtubules, csi3 mutants do not have detectable defects in the velocity of CSCs or the overall alignment of CSCs with microtubules. Furthermore, the inability of ProCSI1:GFP-CSI3 to complement the phenotype of csi1-3 suggests that CSI1 possesses features and functions that are not redundant with CSI3. Although we cannot entirely rule out the possibility that the GFP tag might interfere with the function of this transgenic construct, two similar constructs, ProCSI1:CSI1-GFP and ProCSI3:GFP-CSI3, were able to fully complement csi1 and csi1 csi3 mutants, respectively, indicating that fluorescent protein fusions likely do not disrupt the function of CSI1 or CSI3.
Although csi3 mutants lack an apparent phenotype, the increased severity of csi1-associated phenotypes in the double mutant, csi1 csi3, suggests that CSI3 is involved in some aspects of cellulose biosynthesis. Live-cell imaging was used to more closely investigate the relationship between CSI3, CSI1, and CSCs in vivo. GFP-CSI3 was shown to label two CSC populations, as distinct plasma membrane–localized particles and as isoxaben-induced SmaCCs/MASCs at the cortex of the cell. The high colocalization ratio between CSI3 and CESA6-labeled CSCs was similar to that between CSI1 and CESA6-labeled CSCs. Assuming that the CSCs at the plasma membrane are uniform in composition, one would expect a high colocalization ratio to exist between CSI3 and CSI1 particles as well. Surprisingly, only ∼40% of GFP-CSI3 particles colocalized with RFP-CSI1, which suggests that the composition of CSCs may not be uniform and that CSI1-associated CSCs may rather be segregated from CSI3-associated CSCs. This is consistent with the idea that CSCs are composed of multiple CESA isoforms, some of which can substitute for others. Further support for this hypothesis was garnered from the observation of the temporal behavior of overlapping GFP-CSI3 and RFP-CSI1 signals. Incidents in which overlapping signals were observed were shown to often represent distinct CSCs that were in the process of transiently crossing paths or merging for limited periods of time before splitting into separate particles. SmaCCs/MASCs have been proposed to function as delivery compartments that appear before the insertion of CSCs into the membrane, as storage vesicles of internalized CSCs, or both (Crowell et al., 2009; Gutierrez et al., 2009). The observation that a smaller subset of CSI3 than of CSI1 associated with SmaCCs/MASCs suggests that CSC populations may also be heterogeneous in intracellular SmaCCs/MASCs. Alternatively, CSI1 and CSI3 may associate with different subsets of SmaCCs/MASCs.
Interestingly, in spite of the apparent segregation of CSI1- and CSI3-associated CSCs, there is evidence that suggests each individual CSI protein influences both populations of CSCs. For example, the motility of GFP-CSI3 particles was significantly reduced by the loss of CSI1 in csi1 mutants, suggesting that the motility of CSI3-associated CSCs is dependent on CSI1. This result is consistent with the previous observation that the loss of CSI1 reduced the velocity of YFP-CESA6 (Gu et al., 2010). In addition, the loss of both CSI3 and CSI1 reduced the motility of CSCs in csi1 csi3 by a greater degree than loss of CSI1 alone in csi1, suggesting that both CSI1 and CSI3 influence the normal dynamics of CSCs. Together, the observation that csi1 but not csi3 influences microtubule localization of CSCs and the observation that loss of CSI3 influences CSC velocity in csi1 csi3 suggest that multiple factors influence CSC dynamics.
In 1974, Heath proposed that the interaction between CSCs and microtubules generates a sliding force that moves CSCs within the plasma membrane (Heath, 1974). A few years later, an alternative model dispensed the requirement of microtubules for the motility of CSCs and suggested that glucan polymerization and crystallization propel the CSCs along the membrane (Brown et al., 1976). This alternative hypothesis has been supported by pharmacological and simulation experiments (Robinson and Quader, 1981; Mueller and Brown, 1982; Diotallevi and Mulder, 2007). While there is no requirement for microtubules to make cellulose per se, recent findings suggest that microtubules facilitate the biosynthesis of cellulose microfibrils (Chen et al., 2010; Baskin and Gu, 2012; Li et al., 2012). For instance, microtubule depletion caused by a 10- to 16-h treatment with microtubule-depolymerizing drugs reduces the motility of CSCs. The reduction of CSC motility caused by the pharmacological depletion of microtubules was equivalent to loss of CSI1 in csi1. Since the loss of both CSI1 and CSI3 in csi1 csi3 further reduced the motility of CSCs (Figure 2), it is likely that microtubules influence CSC dynamics along with other factors that may include glucan polymerization and crystallization. The mechanism by which CSI1 and CSI3 influence CSC dynamics may be related to the generation of a gliding force along microtubules. Alternatively, since neither CSI1 nor CSI3 appear to contain a motor domain of any kind, CSI1 and CSI3 may incorporate other non-CESA proteins in the CSC, which may in turn play a role in cellulose synthesis. CSI1 and CSI3 may also play a scaffolding role that is necessary for accommodating proper and efficient cellulose biosynthesis by the CSC. Our data suggest that CSI1 has a major role in the guidance of CSCs along microtubules, whereas CSI3 is dispensable for coalignment of CSCs and microtubules. Loss of CSI1 resulted in loss of association between CSCs and microtubules and reduced CSC motility. The reduction in velocity of CSC in csi1 is likely attributed to a microtubule-dependent function. Together with the observation that removal of microtubules did not affect the distribution and motility of CSCs in csi1, the further reduction of CSC in csi1 csi3 suggests that CSI3 influences the synthesis of cellulose in a microtubule-independent manner. Our data suggest that CSI3 also plays a role in cellulose biosynthesis, but the function of CSI3 is partially dependent on CSI1. In conclusion, we propose that CSI3 influences the velocity of the CSC in both a microtubule-dependent and microtubule-independent manner.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds were surface sterilized using 15% bleach, stratified at 4°C for 3 d, plated on Murashige and Skoog (MS) plates (one-half-strength MS salts, 0.8% agar, and 0.05% MES, pH 5.7), and grown vertically at 22°C in darkness for the specified number of days before drug treatment and imaging. For soil-grown plants, seedlings were germinated and grown on MS plates containing 1% Suc for several days and then transferred to pots in an Arabidopsis growth chamber (Percival) at 22°C under a 16-h-light and 8-h-dark cycle.
Transgenic Lines
GFP-CESA3 and GFP-CESA6 seeds were provided by H. Höfte (Desprez et al., 2007). GFP-CSI3 was constructed in a similar fashion as RFP-CSI1 (Gu et al., 2010). Briefly, a 35S promoter in pH7FWG2 (Karimi et al., 2002) was replaced by a 3-kb CSI3 promoter to create pYG110. The full-length cDNA clone of CSI3 was introduced into pYG111 using Gateway Clonase II (Invitrogen). The verified construct pYG112 was introduced into csi3-1 using Agrobacterium tumefaciens–mediated transformation. RFP-CSI1 plants were constructed as described previously (Gu et al., 2010) and crossed with GFP-CSI3 to create double-labeled transgenic lines. CSI3-RFP was constructed in a similar fashion as GFP-CSI3 except the choice of vector was pH7RWG2. Homozygous YFP-CESA6 prc1-1 seeds (line A6Y-11) were obtained from Chris Somerville (Energy Bioscience Institute, University of California, Berkeley, CA) and crossed with CSI3-RFP to create double-labeled transgenic lines. mCherry-TUA5 seeds were provided by R. Gutierrez and crossed with GFP-CSI3 to create double-labeled transgenic lines. CFP-TUA1 seeds were provided by R. Gutierrez (Gutierrez et al., 2009) and crossed with YFP-CESA6/RFP-CSI3 to generate triple-labeled transgenic lines.
Split-Ubiquitin Yeast Two-Hybrid Assay
The full-length cDNA of CESA3 and CESA6 were PCR amplified and cloned into PCR8 TOPO vector (Invitrogen) using primers in Supplemental Table 1 online. Sequence-confirmed CESA constructs were cloned into CubPLV and NubGX33 (Obrdlik et al., 2004) by Gateway cloning (Invitrogen). The bait expression is regulated by the Met25 promoter, and expression can therefore be regulated by different Met concentrations. The full-length cDNAs of CSI1 and CSI3 were PCR amplified and cloned into the pDONRzeo vector (Invitrogen) using primers in Supplemental Table 1 online. Sequence-confirmed pDONRzeo-CSI1 and pDONRzeo-CSI3 constructs were cloned into CubPLV and NubGX33 by Gateway cloning (Invitrogen).
Saccharomyces cerevisiae strain THY.AP4 was used to cotransform Nub and Cub constructs. NubWT was used as a positive control. NubGX33 was used as a negative control. Cotransformants were selected on synthetic medium lacking Trp and Leu. Cotransformants were grown at 30°C for up to 3 d. For growth assays, cells were grown on synthetic medium lacking Trp, Leu, and His with different concentrations of Met as indicated. The bait was also screened using 15 mM 3-ammonium-triazole in the selection medium to rule out auto activation. β-Galactosidase activity was determined by an in vivo plate assay using X-gal in the medium.
Promoter Swap Lines
The CSI1 promoter was amplified using the primers indicated in Supplemental Table 1 online. Amplified CSI1 promoter was inserted into PCR8 TOPO (Invitrogen). Sequence-confirmed PCR8-proCSI1 was digested using SacI-SpeI and inserted into the pH7FWG2 (Karimi et al., 2002) vector to replace the 35S promoter. The full-length cDNA clone of CSI3 was introduced into pH7FWG2 containing the CSI1 promoter using Gateway Clonase II (Invitrogen). The verified construct pYG113 was introduced into csi1-3 by Agrobacterium-mediated transformation. Transgenic lines were selected on MS medium containing 25 μg/mL of hygromycin and verified by PCR using primers in Supplemental Table 1 online.
Isolation of T-DNA Insertion Line
The identification of the csi3 knockout line from the SIGNAL collection (http://signal.salk.edu/cgi-bin/tdnaexpress) was based on a combination of database searches and PCR amplification of T-DNA flanking regions. For T-DNA lines identified from the SIGNAL collection, seeds were obtained from the ABRC (Ohio State University, Columbus, OH). PCR reactions were performed to identify single plants for the T-DNA insertion. Primers used for T-DNA genotyping of T-DNA mutant alleles are listed in Supplemental Table 1 online.
Cellulose Content Measurement
Crystalline cellulose was measured in 4-d-old etiolated seedlings using the Updegraff method (Updegraff, 1969). Data were collected from five technical replicates for each tissue sample.
Reverse Transcription and PCR Analysis
Total RNA was isolated from Arabidopsis 7-d-old, light-grown seedlings using the RNeasy plant mini kit (Qiagen). For CSI3 and ATC2, 30 cycles of PCR amplification were used using the primers in Supplemental Table 1 online. PCR products were loaded onto 0.8% agarose gels stained with ethidium bromide to visualize the amplified DNAs.
Promoter-GUS Analysis
For GUS construct and staining, genomic DNA fragments (3 kb) upstream from the ATG start codon of CSI2 and CSI3 were cloned into pCAMBIA 1305 GUS-Plus (see Supplemental Table 1 online). The constructs were transformed in to Arabidopsis using Agrobacterium-mediated transformation. Transgenic plants were selected on hygromycin and stained for GUS activity in buffer containing 100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 1 mM ferricyanide, 1 mM ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid at 37°C overnight. Samples were cleared in 70% ethanol and observed under an Olympus Szx7 stereomicroscope.
Confocal Microscopy
Imaging was performed on a Yokogawa CSUX1spinning disk system featuring a DMI6000 Leica motorized microscope, a Photometrics QuantEM:512SC CCD camera, and a Leica ×100/1.4–numerical aperture oil objective. An ATOF laser with three laser lines (440/491/561 nm) was used to enable faster shuttering and switching between different excitations. Band-pass filters (485/30 nm for CFP; 520/50 nm for GFP; 535/30 nm for YFP; 620/60 nm for RFP) were used for emission filtering. Image analysis was performed using Metamorph (Molecular Devices), ImageJ software (version 1.36b; http://rsbweb.nih.gov/ij/), iseeV3.8 (Shenzhen), and Imaris (Bitplane) software.
Drug Treatments
For live-cell imaging, 2-d-old dark-grown seedlings were submerged in MS liquid medium containing the drug and incubated in darkness for various lengths of time. For short-term treatment, 3-d-old dark-grown seedlings were mounted in MS liquid medium containing drugs and imaged at various time points. Oryzalin and isoxaben were dissolved in DMSO to create stock solutions. Stocks were diluted in water directly before each experiment. For mock treatment, seedlings were incubated in appropriately diluted DMSO solution.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At2g22125 (CSI1), At1g44120 (CSI2), At1g77460 (CSI3), At5g05170 (CESA3), and At5g64740 (CESA6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. CSI1-Like Proteins in Arabidopsis.
Supplemental Figure 2. CSI3 Is Expressed in Various Tissues.
Supplemental Figure 3. T-DNA Insertion Analysis of csi3-1 Mutant.
Supplemental Figure 4. Anisotropic Growth Defect and Cellulose Content of the csi1 csi3 Double Mutant.
Supplemental Figure 5. Similar to CSI1, CSI3 Is Localized to Distinct Particles at the Plasma Membrane.
Supplemental Figure 6. CSI3 Partially Associates with SmaCCs/MASCs.
Supplemental Figure 7. Colocalization Analysis among CSI1, CSI3, and Microtubules.
Supplemental Table 1. DNA Primers Used in This Study.
Supplemental Movie 1. Dynamic Association between RFP-CSI1 and GFP-CSI3 (Quantified by Velocity Measurement in Figure 3A).
Supplemental Movie 2. Dynamic Behavior of RFP-CSI1 and GFP-CSI3 (Corresponding to Figures 3E and 3F).
Supplemental Movie 3. Dynamic Association between CSI3 and CESA Complexes.
Supplemental Movie 4. Dynamic Behavior of YFP-CESA6– or RFP-CSI3–Associated SmaCCs/MASCs (Corresponding to Figure 5C).
Supplemental Movie 5. Dynamic Behavior of RFP-CSI1– or GFP-CSI3–Associated SmaCCs/MASCs (Corresponding to Supplemental Figure 7C).
Supplemental Movie 6. Dynamic Association between CSI3 and Microtubules.
Supplemental Movie 7. The CSI3 Velocity Is Dependent on CSI1 (Corresponding to Figure 6F).
Supplemental Movie Legends 1. Brief Descriptions of Supplemental Movies 1 to 7.
Acknowledgments
We thank Ryan Gutierrez and David Ehrhardt for providing mCherryTUA5/YFPCESA6 transgenic seeds and mCherryTUA5 GV3101 agrobacteria strain. We thank Chris Somerville for providing YFPCESA6 and CFPTUA1 transgenic seeds. This work is supported by a grant from the National Science Foundation (1121375).
AUTHOR CONTRIBUTIONS
S.L., L.L., J.D., and Y.G. performed research. S.L., L.L., and Y.G. designed research. S.L., L.L., J.D., L.B., and Y.G. analyzed data. S.L., L.L., L.B., and Y.G. wrote the article.
Glossary
- CESA
cellulose synthase
- CSC
cellulose synthase complex
- SmaCC
small CESA-containing compartment
- MASC
microtubule-associated cellulose synthase compartment
- MS
Murashige and Skoog
References
- Baskin T., Gu Y. (2012). Making parallel lines meet: Transferring information from microtubules to extracellular matrix. Cell Adhes. Migr. 6: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Baskin T.I., Beemster G.T., Judy-March J.E., Marga F. (2004). Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 135: 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M., Li E., Sampathkumar A., Kocabek T., Hauser M.T., Persson S. (2012). POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24: 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.M., Jr, Willison J.H., Richardson C.L. (1976). Cellulose biosynthesis in Acetobacter xylinum: Visualization of the site of synthesis and direct measurement of the in vivo process. Proc. Natl. Acad. Sci. USA 73: 4565–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chen S., Ehrhardt D.W., Somerville C.R. (2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotallevi F., Mulder B. (2007). The cellulose synthase complex: A polymerization driven supramolecular motor. Biophys. J. 92: 2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Cyr R. (2004). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd ed (London: Academic Press), pp. 85–99. [Google Scholar]
- Green P.B. (1962). Mechanism for plant cellular morphogenesis. Science 138: 1404–1405 [DOI] [PubMed] [Google Scholar]
- Gu Y., Kaplinsky N., Bringmann M., Cobb A., Carroll A., Sampathkumar A., Baskin T.I., Persson S., Somerville C.R. (2010). Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Somerville C. (2010). Cellulose synthase interacting protein: A new factor in cellulose synthesis. Plant Signal. Behav. 5: 1571–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Heath I.B. (1974). A unified hypothesis for the role of membrane bound enzyme complexes and microtubules in plant cell wall synthesis. J. Theor. Biol. 48: 445–449 [DOI] [PubMed] [Google Scholar]
- Hepler P., Palevitz B.A. (1974). Microtubules and microfilaments. Annu. Rev. Plant Physiol. 25: 309–362 [Google Scholar]
- Hepler P.K., Newcomb E.H. (1964). Microtubules and fibrils in the cytoplasm of coleus cells undergoing secondary wall deposition. J. Cell Biol. 20: 529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herth W. (1980). Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: Evidence for a gap between polymerization and microfibril formation. J. Cell Biol. 87: 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kimura S., Laosinchai W., Itoh T., Cui X., Linder C.R., Brown R.M., Jr (1999). Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter M.C., Porter K.R. (1963). A “Microtubule” in plant cell fine structure. J. Cell Biol. 19: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., Li S., Gu Y. (2012a). Cellulose synthase complexes: Composition and regulation. Front Plant Sci 3: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., Li S., Gu Y. (2012b). Cellulose synthase interactive protein 1 (CSI1) mediates the intimate relationship between cellulose microfibrils and cortical microtubules. Plant Signal. Behav. 7: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., and Gu, Y. (2012). Cellulose biosynthesis in higher plants and the role of the cytoskeleton. In eLS, A.M. Hetherington, ed (Chichester, UK: John Wiley and Sons), doi/10.1002/9780470015902.a0023745 [Google Scholar]
- Li S., Lei L., Somerville C.R., Gu Y. (2012). Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc. Natl. Acad. Sci. USA 109: 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C. (2011). Dynamic microtubules and the texture of plant cell walls. Int Rev Cell Mol Biol 287: 287–329 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2008). The parallel lives of microtubules and cellulose microfibrils. Curr. Opin. Plant Biol. 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Mueller S.C., Brown R.M., Jr (1982). The control of cellulose microfibril deposition in the cell wall of higher plants : I. Can directed membrane flow orient cellulose microfibrils? Indirect evidence from freeze-fractured plasma membranes of maize and pine seedlings. Planta 154: 489–500 [DOI] [PubMed] [Google Scholar]
- Neville A.C., Gubb D.C., Crawfor R.M. (1976). A new model for cellulose architecture in some plant cell walls. Protoplasma 90: 307–317 [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Robinson D.G., Quader H. (1981). Structure, synthesis, and orientation of microfibrils. IX. A freeze-fracture investigation of the Oocystis plasma membrane after inhibitor treatments. Eur. J. Cell Biol. 25: 278–288 [PubMed] [Google Scholar]
- Roland J.C., Vian B., Reis D. (1975). Observations with cytochemistry and ultracryotomy on the fine structure of the expanding walls in actively elongating plant cells. J. Cell Sci. 19: 239–259 [DOI] [PubMed] [Google Scholar]
- Updegraff D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Wightman R., Turner S.R. (2007). Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 52: 742–751 [DOI] [PubMed] [Google Scholar]