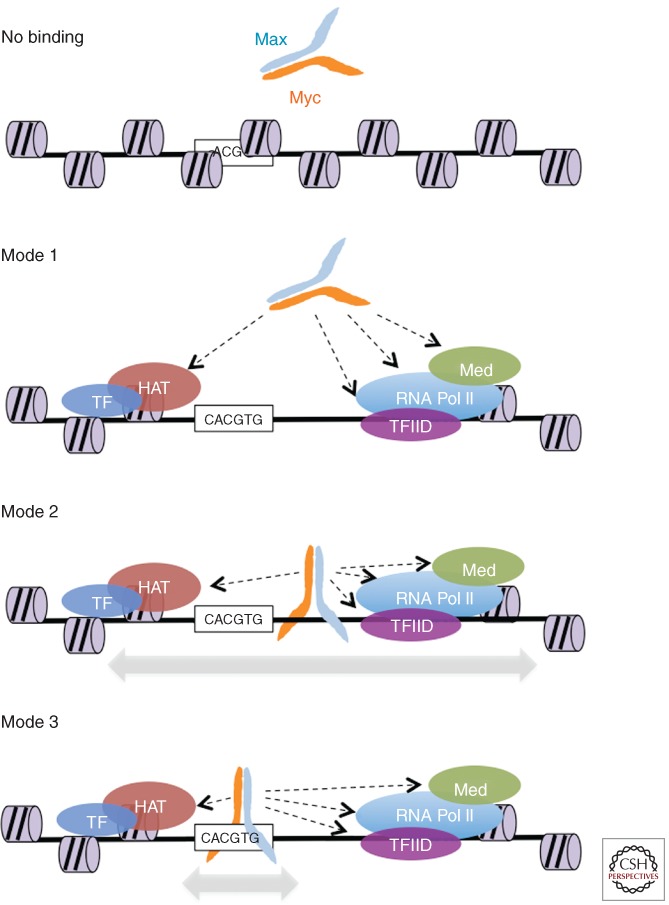
Figure 1.
Different modes of MYC/MAX interaction with DNA. Modes 1–3 not only represent different possible modalities of interaction with genomic sites, but are also postulated to constitute the succession of events by which MYC/MAX dimers are driven to their physiological binding sites (mode 3), as discussed in the text. No binding: access of MYC/MAX to potential sites may be restricted by chromatin or nuclear organization, or by the lack of positive cues for selective recruitment (see text). Indeed, it is important to consider that a large majority of E-box motifs in the genome shows no significant MYC/MAX binding. Mode 1: binding to open chromatin via protein–protein interactions, but without direct DNA contacts. Examples of potentially interacting proteins or protein complexes are schematically represented (HAT, histone acetyltransferase complex; Med, Mediator complex). Mode 2: as mode 1 with additional DNA binding, but in a nonsequence-specific, low-affinity manner. Mode 3: same as above, but stabilized by high-affinity DNA contacts.