Abstract
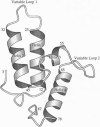
The immunity protein of colicin E7 (ImmE7) can bind specifically to the DNase-type colicin E7 and inhibit its bactericidal activity. Here we report the 1.8-angstrom crystal structure of the ImmE7 protein. This is the first x-ray structure determined in the superfamily of colicin immunity proteins. The ImmE7 protein consists of four antiparallel alpha-helices, folded in a topology similar to the architecture of a four-helix bundle structure. A region rich in acidic residues is identified. This negatively charged area has the greatest variability within the family of DNase-type immunity proteins; thus, it seems likely that this area is involved in specific binding to colicin. Based on structural, genetic, and kinetic data, we suggest that all the DNase-type immunity proteins, as well as colicins, share a "homologous-structural framework" and that specific interaction between a colicin and its cognate immunity protein relies upon how well these two proteins' charged residues match on the interaction surface, thus leading to specific immunity of the colicin.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akutsu A., Masaki H., Ohta T. Molecular structure and immunity specificity of colicin E6, an evolutionary intermediate between E-group colicins and cloacin DF13. J Bacteriol. 1989 Dec;171(12):6430–6436. doi: 10.1128/jb.171.12.6430-6436.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J. U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991 Jul 12;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Bowman C. M., Sidikaro J., Nomura M. Specific inactivation of ribosomes by colicin E3 in vitro and mechanism of immunity in colicinogenic cells. Nat New Biol. 1971 Dec 1;234(48):133–137. doi: 10.1038/newbio234133a0. [DOI] [PubMed] [Google Scholar]
- Chak K. F., Kuo W. S., Lu F. M., James R. Cloning and characterization of the ColE7 plasmid. J Gen Microbiol. 1991 Jan;137(1):91–100. doi: 10.1099/00221287-137-1-91. [DOI] [PubMed] [Google Scholar]
- Cooper P. C., James R. Two new E colicins, E8 and E9, produced by a strain of Escherichia coli. J Gen Microbiol. 1984 Jan;130(1):209–215. doi: 10.1099/00221287-130-1-209. [DOI] [PubMed] [Google Scholar]
- Cowtan K. D., Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr D Biol Crystallogr. 1996 Jan 1;52(Pt 1):43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- Cramer W. A., Dankert J. R., Uratani Y. The membrane channel-forming bacteriocidal protein, colicin El. Biochim Biophys Acta. 1983 Mar 21;737(1):173–193. doi: 10.1016/0304-4157(83)90016-3. [DOI] [PubMed] [Google Scholar]
- Curtis M. D., James R. Investigation of the specificity of the interaction between colicin E9 and its immunity protein by site-directed mutagenesis. Mol Microbiol. 1991 Nov;5(11):2727–2733. doi: 10.1111/j.1365-2958.1991.tb01981.x. [DOI] [PubMed] [Google Scholar]
- Guillet V., Lapthorn A., Hartley R. W., Mauguen Y. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure. 1993 Nov 15;1(3):165–176. doi: 10.1016/0969-2126(93)90018-c. [DOI] [PubMed] [Google Scholar]
- Kossiakoff A. A., Randal M., Guenot J., Eigenbrot C. Variability of conformations at crystal contacts in BPTI represent true low-energy structures: correspondence among lattice packing and molecular dynamics structures. Proteins. 1992 Sep;14(1):65–74. doi: 10.1002/prot.340140108. [DOI] [PubMed] [Google Scholar]
- Ku W. Y., Wang C. S., Chen C. Y., Chak K. F., Safo M. K., Yuan H. S. Crystallization and preliminary X-ray crystallographic analysis of ImmE7 protein of colicin E7. Proteins. 1995 Dec;23(4):588–590. doi: 10.1002/prot.340230414. [DOI] [PubMed] [Google Scholar]
- Ohno-Iwashita Y., Imahori K. Assignment of the functional loci in colicin E2 and E3 molecules by the characterization of their proteolytic fragments. Biochemistry. 1980 Feb 19;19(4):652–659. doi: 10.1021/bi00545a008. [DOI] [PubMed] [Google Scholar]
- Ohno-Iwashita Y., Imahori K. Assignment of the functional loci in the colicin E1 molecule by characterization of its proteolytic fragments. J Biol Chem. 1982 Jun 10;257(11):6446–6451. [PubMed] [Google Scholar]
- Osborne M. J., Lian L. Y., Wallis R., Reilly A., James R., Kleanthous C., Moore G. R. Sequential assignments and identification of secondary structure elements of the colicin E9 immunity protein in solution by homonuclear and heteronuclear NMR. Biochemistry. 1994 Oct 18;33(41):12347–12355. doi: 10.1021/bi00207a001. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P. The ins and outs of colicins. Part I: Production, and translocation across membranes. Microbiol Sci. 1984 Oct;1(7):168–175. [PubMed] [Google Scholar]
- Schaller K., Nomura M. Colicin E2 is DNA endonuclease. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3989–3993. doi: 10.1073/pnas.73.11.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs M. T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990 Jun;9(6):1939–1947. doi: 10.1002/j.1460-2075.1990.tb08321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi D. M., Obendorf S. K., Moffat K. Structure of vitamin D-dependent calcium-binding protein from bovine intestine. Nature. 1981 Nov 26;294(5839):327–332. doi: 10.1038/294327a0. [DOI] [PubMed] [Google Scholar]
- Toba M., Masaki H., Ohta T. Colicin E8, a DNase which indicates an evolutionary relationship between colicins E2 and E3. J Bacteriol. 1988 Jul;170(7):3237–3242. doi: 10.1128/jb.170.7.3237-3242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis R., Leung K. Y., Pommer A. J., Videler H., Moore G. R., James R., Kleanthous C. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the millimolar to femtomolar affinity range. Biochemistry. 1995 Oct 24;34(42):13751–13759. doi: 10.1021/bi00042a005. [DOI] [PubMed] [Google Scholar]
- Wallis R., Moore G. R., Kleanthous C., James R. Molecular analysis of the protein-protein interaction between the E9 immunity protein and colicin E9. Eur J Biochem. 1992 Dec 15;210(3):923–930. doi: 10.1111/j.1432-1033.1992.tb17496.x. [DOI] [PubMed] [Google Scholar]
- Wallis R., Reilly A., Barnes K., Abell C., Campbell D. G., Moore G. R., James R., Kleanthous C. Tandem overproduction and characterisation of the nuclease domain of colicin E9 and its cognate inhibitor protein Im9. Eur J Biochem. 1994 Mar 1;220(2):447–454. doi: 10.1111/j.1432-1033.1994.tb18642.x. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]